Background and Aims:
HBV RNA in peripheral blood reflects HBV cccDNA transcriptional activity and may predict clinical outcomes. The prospective Melbourne HBV-STOP trial studied nucleot(s)ide analog discontinuation in HBeAg-negative non-cirrhotic participants with long-term virological suppression. Ninety-six weeks after stopping treatment, the proportion of participants with virological relapse (HBV DNA > 2000 IU/mL), biochemical relapse (ALT > 2 × ULN and HBV DNA > 2000 IU/mL), or hepatitis flare (ALT > 5 × ULN and HBV DNA > 2000 IU/mL) was 89%, 58%, and 38%, respectively. We evaluated the ability of serum HBV RNA levels to predict these outcomes.
Approach & Results:
HBV RNA levels were measured using the Roche cobas 6800/8800 HBV RNA Investigational Assay. Sixty-five participants had baseline and longitudinal off-treatment specimens available for RNA testing. HBV RNA was detectable at baseline in 25% of participants and was associated with a higher risk of biochemical relapse (81% vs. 51%, p value 0.04) and hepatitis flare (63% vs. 31%, p value 0.04). Participants who had undetectable serum HBV RNA as well as HBsAg ≤ 100 IU/mL at baseline were less likely to experience virological relapse (4 of 9, 44%) than participants with detectable HBV RNA and HBsAg level > 100 IU/mL (15/15, 100%; p value 0.0009). Off-treatment levels of HBV RNA were correlated with HBV DNA and were associated with the risk of hepatitis flare.
Conclusions:
Serum HBV RNA may be a useful biomarker for guiding clinical decision-making before stopping nucleot(s)ide analog therapy. Baseline HBV RNA and HBsAg levels are associated with the risk of clinical relapse, hepatitis flare, and disease remission off-treatment.
INTRODUCTION
Nucleot(s)ide analogs (NA) are the standard treatment for chronic hepatitis B (CHB) infection.1–3 However, HBsAg clearance, or functional cure, is rare and long-term treatment is recommended. Finite therapy for individuals with HBeAg-negative CHB has been identified as an area of unmet need, and in the past decade, there has been increasing interest in whether NA therapy can be safely stopped in a subset of participants.4–6 The amount of circulating HBV RNA in peripheral blood may be a marker of HBV covalently closed circular DNA (cccDNA) transcriptional activity within infected hepatocytes and predict clinical outcomes. We recently published the results of a large, prospective cohort study evaluating treatment withdrawal in a cohort of non-cirrhotic participants with HBeAg-negative CHB.7 After 96 weeks of follow-up, 6% of participants had lost HBsAg, 28% were in disease remission (HBV DNA < 2000 IU/mL and ALT < 2 × ULN), 30% had restarted NA therapy, and hepatitis flare (ALT > 5 × ULN) occurred in 32%.7 However, the results of clinical trials evaluating outcomes after stopping NA therapy have been heterogenous,4,8–17 with different rates of virological relapse, hepatitis flare, and HBsAg seroclearance impacted by study design. There is a need for biomarkers that predict clinical outcomes more accurately after stopping NA therapy.
A low level of HBsAg in serum before cessation of NA therapy is associated with the maintenance of viral suppression, with most studies applying a threshold of 100 IU/mL.8,12,16,18 In our prospective study, serum HBsAg levels below 10 IU/mL at baseline were associated with HBsAg loss as well as the likelihood of disease remission.7 A second potential biomarker is HBV RNA, an important intermediate in the HBV life cycle. Transcription of cccDNA, the nuclear mini-chromosome of HBV, forms 5 viral RNA transcripts. The RNA transcripts are translated to form the HBV proteins, as well as forming the replicative template for reverse transcriptase to DNA. Pre-genomic RNA (pgRNA), the second largest RNA transcript at 3.5kb, is encapsidated in the cytoplasm before reverse transcription to DNA, assembly to form mature virions, and secretion from the hepatocyte. Encapsidated RNA may be detected in peripheral blood in addition to the HBV DNA found in mature virions.19 Emerging evidence suggests that serum HBV RNA negativity may predict the risk of virological relapse after stopping NA therapy, although to date, results have been limited by both study design and the use of variable in-house assays.8,20–22
Early assays for estimating HBV RNA were indirect and based on the subtraction of the HBV DNA component from the measurement of total nucleic acid.21 Subsequent assays for detection of peripheral HBV RNA have been designed using rapid amplification of cDNA Ends (20–22 reviewed in23). These in-house assays, which utilize the polyadenylated tail of RNA, are labor-intensive and can lack sensitivity. Furthermore, as there is no international standard, assays are not directly comparable unless communal standards are employed. Recently, Roche has developed an automated assay on the cobas 6800/8800 platforms for the determination of HBV RNA levels in serum or plasma.19,24,25
Therefore, the aim of this study was to evaluate the ability of serum HBV RNA levels to predict clinical outcomes after stopping NA therapy in a large cohort of non-cirrhotic individuals with HBeAg-negative CHB enrolled in a prospective cohort trial.
METHODS
Participants and outcome variables
We analyzed specimens from an investigator-initiated, prospective, multicenter, observational cohort study of the clinical outcomes after stopping NA therapy among non-cirrhotic people with HBeAg-negative CHB and sustained virological suppression (undetectable serum HBV DNA) on-treatment.7 The study was approved by the St Vincent’s Hospital Ethics Committee (ID Number: 032/14) and at each participating site and was conducted in compliance with the principles of the 1975 Declaration of Helsinki and local regulatory requirements. All participants provided informed consent before screening. Participants were aged over 18 years, with non-cirrhotic HBeAg-negative CHB at the time of NA treatment initiation and were followed for 96 weeks. The key inclusion criteria were: age > 18 years; HBeAg-negative CHB at the time NA therapy was started; continuous NA therapy for at least 2 years with HBV DNA levels < 20 IU/mL on 3 consecutive occasions at least 6 months apart over 18 months; and the absence of cirrhosis (liver stiffness ≤ 9.5 kPa at screening, and if available, liver histology showing METAVIR stage ≤F3 before starting NA therapy).7
Sixty-five participants had serum specimens stored at -80°C from baseline (the day that NA treatment was stopped) and longitudinal off-treatment time points. We considered the ability of HBV RNA levels to predict the following clinical outcomes after stopping NA: (i) virological relapse (HBV DNA > 2000 IU/mL); (ii) biochemical relapse (ALT >2 × ULN and HBV DNA >2000 IU/mL); (iii) hepatitis flare (ALT >5 × ULN and HBV DNA >2000 IU/mL); and (iv) HBsAg loss.
Laboratory tests
HBV DNA and HBsAg in serum were measured prospectively during the clinical study. Serum HBV DNA was measured using the cobas AmpliPrep/COBAS TaqMan platform (Roche Diagnostics), which has a lower limit of detection (LOD) of 20 IU/mL. Results below the LOD were assigned an arbitrary value of zero for analysis. Serum HBsAg was measured using the Elecsys HBsAg II quant II (Roche Diagnostics) with a LOD of 0.05 IU/mL. Serum HBV RNA levels were measured retrospectively on stored samples using the Roche cobas 6800/8800 investigational HBV RNA assay (Roche Molecular Systems, CA, US). The cobas 6800/8800 investigational HBV RNA assay is a high throughput, sensitive and inclusive assay that has been developed and validated as a tool to evaluate the clinical relevance of HBV RNA quantification in participants with CHB. The amplification target is located at the 3′ end of HBV transcripts, enabling it to detect all HBV RNAs expressed from cccDNA. Assay specificity for RNA versus DNA is enhanced with a blocking oligonucleotide that binds to the HBV DNA genome in competition with an assay primer, as reported.19 This investigational assay has an LOD of 5 copies/mL, a lower limit of quantitation of 10 copies/mL, and a linear range of 10–109 copies/mL. Results below the LOD were assigned an arbitrary value of zero, and results with detectable target below the lower limit of quantitation were assigned an arbitrary value of 5 copies/mL.
Statistical analysis
Statistical comparisons were carried out using the Wilcoxon Rank-Sum test for continuous variables and χ2 test or Fisher’s Exact test for categorical variables with the Freeman-Halton26 extension for comparison across more than 2 categories. The proportion difference test was also used for additional post hoc analysis. The cumulative rate of clinical outcomes was calculated using the cumulative incidence functions method, with Gray’s test for equality27 used for comparisons where appropriate. Threshold values used to delineate discrete time periods were assigned at times when the curves cross, since in this case, the proportionality assumption for the hazard functions fails, and the log-rank test is no longer appropriate. Therefore, the p values were calculated based on the separate regions. Data were right-censored when a subject left the study before an event occurred or when the study ended before the event had occurred. Receiver operating characteristic (ROC) curve analysis was used for optimal threshold identification. A combination of ROC curve cutoff metrics (Youden index, distance to (0,1) and the absolute value of the sensitivity and specificity difference) was employed to search for candidate threshold level optima.28 The optimal threshold was defined as the midpoint between the 2 measured values with the highest Youden indices. All data were analyzed using SAS System software Release 9.4 (The SAS Institute, Cary, NC, USA). A 2-tailed p value less than 0.05 was considered as statistically significant.
RESULTS
Participant characteristics at baseline
Sixty-five participants had serum available for RNA testing at baseline and longitudinally off-treatment. There were between 3 and 18 (median: 10) follow-up samples collected per participant, collected at times from stopping therapy ranging between 18 and 672 days (total number of samples: 655). The baseline characteristics of the cohort are presented in Table 1. At the time of stopping NA therapy, the median age was 56 (range: 31–74) years, 35 (54%) were male, and 49 (75%) were Asian. The median HBsAg level was 703 (range: 1.6–19,912) IU/mL. Baseline NA treatment was entecavir (ETV) for 42 (65%) participants, tenofovir (TDF) for 17 (26%) participants, lamivudine for 3 (4.6%) participants, and lamivudine plus adefovir for 3 (4.6%) participants.
TABLE 1.
Participant characteristics at baseline
| Characteristic | N (%) or median (range) |
|---|---|
| Male gender (n, %) | 35 (54) |
| Age, y (median, range) | 56 (31–74) |
| Ethnicity (n, %) | |
| Asian | 49 (75) |
| Caucasian | 7 (11) |
| Other | 9 (14) |
| Serum ALT, IU/mL (median, range) | 24 (8–60) |
| Serum HBsAg level, IU/mL (median, range) | 701 (1.6–19,912) |
| Serum HBsAg level < 100 IU/mL (n, ) | 10 (15) |
| Serum HBsAg level <10 IU/mL (n, %) | 7 (11) |
| NA therapy at baseline (n, %) | |
| Entecavir | 42 (65) |
| Tenofovir | 17 (26) |
| Lamivudine | 3 (4.6) |
| Lamivudine/adefovir | 3 (4.6) |
| HBV genotype (n, %)a | |
| A | 3 (5) |
| B | 28 (43) |
| C | 14 (22) |
| D | 10 (15) |
| E | 1 (2) |
| unknown | 9 (14) |
Inferred baseline genotype, based on sequencing of HBV DNA before becoming undetectable or after virological relapse.
Abbreviations: ALT, alanine aminotransferase; NA, nucleot(s)ide analogs.
At the time of stopping NA, serum HBV RNA was detectable in 16 (25%) participants. Among participants with detectable HBV RNA, the median serum HBV RNA level was 13 copies/mL (range < 10–62 copies/mL). No baseline participant characteristic or viral biomarker measured (age, gender, ethnicity, NA used , that is, ETV vs. TDF, ALT, HBsAg level) was significantly associated with detectable serum HBV RNA.
Factors associated with biochemical relapse and hepatitis flare after stopping NA therapy
Following treatment withdrawal, virological relapse (HBV DNA > 2000 IU/mL) was observed in 58 (89%) participants by week 96. Biochemical relapse (ALT > 2 × ULN and HBV DNA >2000 IU/mL) occurred in 38 (58%) participants, hepatitis flare (ALT > 5 × ULN and HBV DNA >2000 IU/mL) in 25 (38%) and HBsAg loss in 4 (6%). NA therapy was restarted in 17 (26%) participants. Stopping NA therapy was safe and well-tolerated, as reported.7
Detectable serum HBV RNA at baseline was associated with a higher risk of biochemical relapse (13/16, 81% vs. 25/49, 51%, p value 0.043; Fig. 1A and Table S1, http://links.lww.com/HC9/A340) as well as a higher risk of hepatitis flare by week 96 (10/16, 63% vs. 15/49, 31%, p value 0.037; Fig. 1A and Table S1, http://links.lww.com/HC9/A340). The detection of serum HBV RNA at baseline was not significantly associated with the likelihood of virological relapse by week 96 (16/16 (100%) vs. 42/49 (86%), p value 0.18). Cumulative incidence plots for time to virological relapse, biochemical relapse, and hepatitis flare in participants with detectable versus undetectable HBV RNA at baseline are shown in Supplemental Data Figure S1, http://links.lww.com/HC9/A340. No participant with detectable serum HBV RNA at baseline achieved HBsAg loss, compared to 8% of participants with undetectable HBV RNA (p value = 0.56). Six of 16 participants (38%) with detectable HBV RNA in serum were restarted on NA therapy, compared to 11 of 49 (22%) participants with undetectable HBV RNA (p value = 0.3).
FIGURE 1.
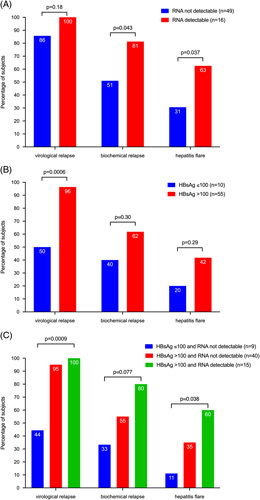
Proportions of participants with virological relapse (HBV DNA > 2000 IU/mL), biochemical relapse (ALT > 2 × ULN and HBV DNA >2000 IU/mL), and hepatitis flare (ALT >5 × ULN and HBV DNA >2000 IU/mL) at week 96 according to baseline HBV RNA detection and HBsAg level. (A) Association with baseline RNA detectability. (B) Association with HBsAg level (above or below 100 IU/mL). (C) Association with the combination of RNA detectability and HBsAg level. One participant (not shown) with HBsAg ≤ 100 IU/mL and detectable HBV RNA had virological and biochemical relapse and hepatitis flare. p values from Fisher’s Exact Test for HBV RNA and HBsAg, or Freeman-Halton26 for the combination. Abbreviations: ALT, alanine aminotransferase; ULN, upper limit of normal.
HBsAg levels >100 IU/mL have previously been associated with the risk of virological relapse after stopping NA therapy, suggesting that this threshold can be used to guide the consideration of treatment cessation.8 In our study, virological relapse was more common in participants with baseline HBsAg level >100 IU/mL (53/55, 96%) than in those with baseline HBsAg ≤ 100 IU/mL (5/10, 50%; p value 0.0006, Fig. 1B). There was no significant association between baseline HBsAg > 100 IU/mL and biochemical relapse or hepatitis flare (Fig. 1B and Table S1, http://links.lww.com/HC9/A340). We also tested the association between HBsAg > 1000 IU/mL and clinical outcomes but found no significant association between HBsAg > 1000 IU/mL and risk of virological relapse, biochemical relapse, or hepatitis flare (Supplemental Table S1, http://links.lww.com/HC9/A340).
The combination of serum HBV RNA detectability and HBsAg level (<100 IU/mL vs. ≥100 IU/mL) at baseline provided additional clinical information. Participants who had undetectable serum HBV RNA as well as HBsAg ≤100 IU/mL at baseline were less likely to experience virological relapse (4 of 9, 44%) than participants with detectable HBV RNA and HBsAg level >100 IU/mL (15/15, 100%; p value 0.0009; Fig. 1C). Similarly, the risk of hepatitis flare was significantly higher in participants with detectable HBV RNA and HBsAg level >100 IU/mL at baseline (9/15, 60% vs. 1/9, 11%; p value 0.038).
Serum HBV RNA levels after stopping NA
Among participants who were negative for serum HBV RNA at baseline, HBV RNA became detectable in 46 of 49 participants (94%) by week 96 of follow-up (Supplemental Data Figure S2, http://links.lww.com/HC9/A340). The median time to detection of HBV RNA was 126 days (range: 26–672 days). The time to detection of serum HBV RNA was significantly shorter for participants stopping TDF (median 55 days, range 26–672) versus ETV (median 133 days, range 28–672; Mann-Whitney p value 0.0011, Figure S2, http://links.lww.com/HC9/A340). However, the difference between the cumulative incidence of RNA detectability by week 96 in patients who had been treated with TDF and those who had been treated with ETV was not statistically significant (Gray’s test for equality p = 0.12; Figure S2, http://links.lww.com/HC9/A340).
HBV RNA and DNA levels in serum following cessation of NA therapy were well correlated with each other (Fig. 2). Among post-baseline samples, 346 had both detectable RNA and DNA, 98 had both undetectable, 190 had DNA detectable, but RNA undetectable, and 19 had DNA undetectable but RNA detectable. Fluctuations in mean serum HBV RNA and DNA concentrations paralleled each other over time (Supplemental Data Figure S3, http://links.lww.com/HC9/A340).
FIGURE 2.
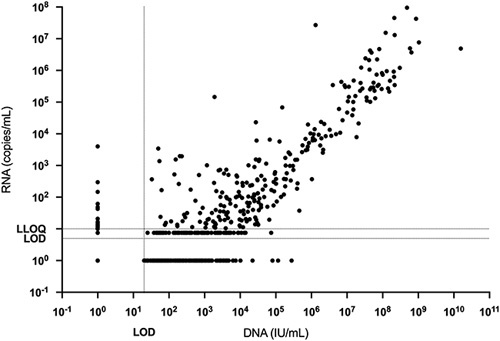
Correlation between serum HBV DNA and RNA in participants following the cessation of NA therapy. Results below the LOD or between the LOD and LLOQ were assigned nominal values as described in the Methods for illustrative purposes. Abbreviations: LLOQ, lower limit of quantitation; LOD, limit of detection; NA, nucleot(s)ide analogs.
We observed a statistically significant difference in post-baseline peak serum HBV RNA level between participants who experienced a hepatitis flare (median 5.0 log10 copies/mL, range 1.6–8.0 log10 copies/mL) compared to those who did not (median 1.3 log10 copies/mL, range undetectable to 3.6 log10 copies/mL; p value < 0.0001, Wilcoxon Rank-Sum test).
Prognostic value of serum RNA or DNA concentration for prediction of hepatitis flare
The observation that RNA concentrations reached higher levels in participants who experienced a hepatitis flare suggests the possibility that an HBV RNA increase above a certain threshold might serve as a prognostic warning indicator of hepatitis flare. We assessed the HBV RNA concentration at the time point before the hepatitis flare, which was a median of 28 days (range: 14–84 days) before the flare was detected. The median serum HBV RNA level at this time point was 2.65 log10 copies/mL (range: undetectable to 6.67 log10 copies/mL), which was significantly higher than the median peak HBV RNA level in participants without flare (p value <0.05). The optimal HBV RNA threshold for prediction of a hepatitis flare at the subsequent time point was 128 copies/mL (area under ROC curve and 95% confidence interval: 0.67 (0.56–0.79); Supplemental Data Figure S4, http://links.lww.com/HC9/A340). A similar analysis was also performed using HBV DNA; the optimal threshold was 16,637 IU/mL (area under ROC curve and 95% CI: 0.70 (0.60–0.80). The positive percent agreement (58 vs. 63%), negative percent agreement (77% for both), and overall percent agreement (76% for both) were similar for RNA and DNA using these optimized thresholds (95% CIs overlap—see Table S2, http://links.lww.com/HC9/A340).
Biomarker profiles in individual patients, illustrating the relationship between serum HBV RNA and DNA with subsequent ALT flares, are shown in Supplemental Data Figure S5, http://links.lww.com/HC9/A340.
DISCUSSION
Several effective and well-tolerated NA drugs are available for the treatment of HBV infection. However, there is great interest in determining whether NA therapy can be stopped in a subset of people with HBeAg-negative CHB who have achieved long-term viral suppression, followed by maintenance of long-term viral suppression or even the achievement of HBsAg seroclearance.4,29,30 However, the prediction of which patients are more likely to achieve these outcomes remains challenging. Low HBsAg levels have been proposed as a useful biomarker,7,9,31 but there is a need for other biomarkers with better clinical utility.
The detection and quantification of circulating HBV RNA in blood have recently emerged as a potential biomarker for predicting clinical outcomes in CHB.8,22,32–35 The present study demonstrated, in a cohort of non-cirrhotic participants with HBeAg-negative CHB who were prospectively recruited to an observational cohort study, the clinical value of serum HBV RNA measurements for determining suitability for stopping NA therapy. Serum HBV RNA was detectable in 25% of participants at baseline and was associated with an increased risk of biochemical relapse and hepatitis flare. None of the participants with detectable serum HBV RNA at the time of stopping NA achieved HBsAg loss after 96 weeks of follow-up. The presence of HBV RNA in serum at baseline therefore helps to identify participants who should not be considered for stopping NA therapy.
The composite variable of HBsAg <100 IU/mL and undetectable HBV RNA provided additional information compared to either marker alone and was useful for identifying the subset of participants with a lower risk of virological relapse, biochemical relapse, and hepatitis flare. Participants with HBsAg levels > 100 IU/mL but undetectable HBV RNA had an intermediate risk of biochemical relapse or hepatitis flare. Future prospective studies will be required to confirm optimal thresholds for clinical use.
HBV RNA became detectable in 94% of participants after NA therapy was stopped, reflecting increased HBV replication. The shorter time to RNA detectability in participants who had been treated with TDF versus ETV is consistent with our previous observation regarding serum DNA levels.7 The mechanism underlying the difference in the rate of relapse between participants stopping TDF versus ETV remains unexplained, and the long-term clinical relevance of the observation is not clear. It may be a consideration for the design of future clinical protocols that include stopping rules for NA therapy with short-term follow-up.
Rising HBV RNA levels off-treatment were associated with hepatitis flares; HBV RNA above 23–32 copies/mL off-treatment was a threshold associated with hepatitis flare at a later time point. Off-treatment serum HBV RNA levels correlated strongly with HBV DNA levels, so it was not possible to statistically test whether RNA or DNA levels were more useful for predicting hepatitis flares. We note that quantitative HBV DNA, though not RNA, assays are routinely available in clinical practice.
Our data confirm and extend previous studies of serum HBV RNA measurement as a biomarker for participants being considered for withdrawal from NA therapy. Laras et al36 recently reported an association between serum pgRNA and clinical outcomes after stopping NA in a retrospective cohort of 74 Greek participants with HBeAg-negative CHB. pgRNA was only detectable in 7/74 (9.5%) of participants at baseline; these participants were highly likely to experience virological and clinical relapse, and none lost HBsAg. A high rate of HBsAg loss was observed in pgRNA-negative participants (49% at a median of 12 months). Kaewdech and colleagues observed similar findings in a Thai cohort of mixed HBeAg-positive and HBeAg-negative CHB who met the APASL criteria for stopping NA and were followed for 48 weeks. Serum HBV RNA at baseline predicted the risk of virological and clinical relapse. Longitudinal samples off-treatment were not tested in either of these studies. Seto and colleagues evaluated the use of serum HBV RNA and HBsAg measurements in stratifying off-treatment relapse risk in a selected cohort of Asian participants with mixed HBeAg-positive and HBeAg-negative CHB who all had low HBsAg levels (< 200 IU/mL), using the Abbott m2000 platform.37 They found the combination of HBsAg levels < 10 IU/mL and undetectable HBV RNA to be associated with a lower risk of virological relapse, but due to the study design, there was limited power to test the use of HBV RNA as a biomarker in participants with higher HBsAg levels, or the association between HBV RNA and biochemical relapse or hepatitis flare.
The association between baseline serum HBV RNA levels and the risk of biochemical relapse and hepatitis flare off-treatment most likely reflects the transcriptional activity of the intranuclear HBV cccDNA, which is the template for reverse transcription, the step in the viral life cycle targeted by NA. Serum HBV RNA has been shown to correlate with intrahepatic pgRNA and to reflect continued cccDNA transcription during NA therapy in the absence of detectable serum HBV DNA.38 When NA is withdrawn, HBV replication rapidly increases, driving the risk of hepatitis.
There are several possible explanations for the observed increase in HBV RNA levels after NA cessation. Nuclear cccDNA levels may be increased as a result of increased viral replication activity and associated intranuclear recycling of nascent capsids. It is also possible that the transcriptional activity of cccDNA increases as an indirect consequence of increased production of viral proteins including HBx. The HBx protein can enhance HBV replication by promoting the degradation of the “structural maintenance of chromosomes” (Smc) complex Smc5/6.39 In the absence of HBx, active Smc5/6 serves as a restriction factor that blocks the transcription from extrachromosomal HBV DNA. HBx also affects the recruitment of histone acetyltransferases such as HAT-1 that regulate the acetylation status of cccDNA-bound histone and of coactivators involved in chromatin modulation, both pathways by which cccDNA transcriptional activity may increase.40–48 NA cessation studies provide a novel opportunity for translational studies of this phenomenon and possibly a deeper understanding of the regulatory mechanisms involved that may have implications for novel therapeutic development.
This study has several limitations. HBsAg loss at 96 weeks was infrequent, limiting the power to detect an association between HBV RNA detection and HBsAg loss. This is typical of more recent studies that have recruited participants prospectively, avoiding the potential bias that may occur with retrospective data collection. Nonetheless, it was notable that no participants with detectable serum HBV RNA at baseline achieved HBsAg loss. We designed this study to specifically evaluate the role of serum HBV RNA as a biomarker in participants stopping NA therapy; we did not consider other novel biomarkers, such as hepatitis B core-related antigen. Recent data have been mixed regarding the potential association between hepatitis B core-related antigen and clinical outcomes.8,49 Further studies will be needed to establish the clinical role of this biomarker; the assay can be difficult to access.
In conclusion, measurement of serum HBV RNA, in combination with other biomarkers, may be useful for the triage of patients with HBeAg-negative CHB and undetectable serum HBV DNA who are considering stopping NA therapy. Participants with detectable HBV RNA have a higher risk of biochemical relapse and hepatitis flare and are unlikely to achieve HBsAg loss. The combination of undetectable HBV RNA and low HBsAg levels at baseline can identify patients who are most suitable for stopping NA therapy, with a lower risk of virological relapse, clinical relapse, and hepatitis flare. Additional studies to confirm these observations are warranted.
Supplementary Material
Acknowledgments
ACKNOWLEDGMENTS
The authors thank the patients who participated in the study. Manuscript preparation was supported by Data First Consulting, Inc. (Sebastopol, CA) and funded by Roche Molecular Systems.
FUNDING INFORMATION
The clinical study was supported by the National Health and Medical Research Council of Australia Project Grant 1066536. Additional support for data extraction was provided by Roche Molecular Systems, Inc. Alexander Thompson received funding from the National Health and Medical Research Council of Australia (MRFF Practitioner Fellowship 1142976). cobas® 6800/8800 HBV RNA Investigational Assay reagents were provided by Roche Molecular Systems, Inc.
CONFLICTS OF INTEREST
Amanda J. Nicoll advises and is on the speakers’ bureau for Ipsen, Eisai, Roche, and AstraZeneca. Simone I. Strasser advises and is on the speakers’ bureau for Gilead, Eisai, Norgine, Roche, Chiesi, and AstraZeneca. She advises Pfizer and MSD and is on the speakers’ bureau for Otsuka and AbbVie. Gail Matthews advises and received grants from Gilead and ViiV. She advises AstraZeneca and received grants from AbbVie. Kumar Visvanathan consults, advises, and is on the speakers’ bureau for Gilead. He advises ViiV. The remaining authors have no conflicts to report.
Footnotes
Abbreviations: ALT, alanine aminotransferase; cccDNA, covalently closed circular DNA; CHB, chronic hepatitis B; ETV, entecavir; LLOQ, lower limit of quantitation; LOD, limit of detection; NA, nucleot(s)ide analog; pgRNA, pre-genomic RNA; ROC, receiver operating characteristic; TDF, tenofovir disproxil fumarate; ULN, upper limit of normal.
Supplemental Digital Content is available for this article. Direct URL citations are provided in the HTML and PDF versions of this article on the journal’s website, www.hepcommjournal.com.
Contributor Information
Alexander J. Thompson, Email: alexander.thompson@svha.org.au.
Kathy Jackson, Email: kathy.jackson@vidrl.org.au.
Sara Bonanzinga, Email: Sara.Bonanzinga@vidrl.org.au.
Sam A.L. Hall, Email: sammy.hall86@gmail.com.
Simon Hume, Email: simon.hume@svha.org.au.
Gareth S. Burns, Email: garethburns@gmail.com.
Vijaya Sundararajan, Email: vijaya.sundararajan@unimelb.edu.au.
Dilip Ratnam, Email: Dilip.Ratnam@monash.edu.
Miriam T. Levy, Email: Miriam.Levy@health.nsw.gov.au.
John Lubel, Email: johnlubel@icloud.com.
Amanda J. Nicoll, Email: amanda.nicoll@easternhealth.org.au.
Simone I. Strasser, Email: simone.strasser@health.nsw.gov.au.
William Sievert, Email: william.sievert@monash.edu.
Paul V. Desmond, Email: paul.desmond@svha.org.au.
Meng C. Ngu, Email: Meng.Ngu@sswahs.nsw.gov.au.
Marie Sinclair, Email: marie.sinclair@austin.org.au.
Christopher Meredith, Email: drcmeredith@gmail.com.
Gail Matthews, Email: gmatthews@kirby.unsw.edu.au.
Peter A. Revill, Email: Peter.Revill@vidrl.org.au.
Margaret Littlejohn, Email: margaret.littlejohn@vidrl.org.au.
D. Scott Bowden, Email: scott.bowden@vidrl.org.au.
Jesse A. Canchola, Email: jesse.canchola@roche.com.
Jason Torres, Email: jason.torres@roche.com.
Philip Siew, Email: philip.siew@roche.com.
Jasmin Lau, Email: jasmine.lau@roche.com.
Alison Kuchta, Email: alison.kuchta@roche.com.
Kumar Visvanathan, Email: kv@unimelb.edu.au.
REFERENCES
- 1. European Association for the Study of the Liver. EASL 2017 Clinical Practice Guidelines on the management of hepatitis B virus infection. J Hepatol. 2017;67:370–398. [DOI] [PubMed] [Google Scholar]
- 2. Terrault NA, Lok ASF, McMahon BJ, Chang KM, Hwang JP, Jonas MM, et al. Update on prevention, diagnosis, and treatment of chronic hepatitis B: AASLD 2018 hepatitis B guidance. Hepatology. 2018;67:1560–1599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. World Health Organization. Updated recommendations on simplified service delivery and disagnostics for hepititis C infection: policy brief. In; 2022.
- 4. Hall SAL, Vogrin S, Wawryk O, Burns GS, Visvanathan K, Sundararajan V, et al. Discontinuation of nucleot(s)ide analogue therapy in HBeAg-negative chronic hepatitis B: a meta-analysis. Gut. 2022;71:1629–1641. [DOI] [PubMed] [Google Scholar]
- 5. van Bommel F, Berg T. Risks and benefits of discontinuation of nucleos(t)ide analogue treatment: A treatment concept for patients with HBeAg-negative chronic hepatitis B. Hepatol Commun. 2021;5:1632–1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kao JH, Jeng WJ, Ning Q, Su TH, Tseng TC, Ueno Y, et al. APASL guidance on stopping nucleos(t)ide analogues in chronic hepatitis B patients. Hepatol Int. 2021;15:833–851. [DOI] [PubMed] [Google Scholar]
- 7. Hall SAL, Burns GS, Anagnostou D, Vogrin S, Sundararajan V, Ratnam D, et al. Stopping nucleot(s)ide analogues in non-cirrhotic HBeAg-negative chronic hepatitis B patients: HBsAg loss at 96 weeks is associated with low baseline HBsAg levels. Aliment Pharmacol Ther. 2022;56:310–320. [DOI] [PubMed] [Google Scholar]
- 8. Seto WK, Liu KS, Mak LY, Cloherty G, Wong DK, Gersch J, et al. Role of serum HBV RNA and hepatitis B surface antigen levels in identifying Asian patients with chronic hepatitis B suitable for entecavir cessation. Gut. 2021;70:775–783. [DOI] [PubMed] [Google Scholar]
- 9. Sonneveld MJ, Chiu SM, Park JY, Brakenhoff SM, Kaewdech A, Seto WK, et al. Probability of HBsAg loss after nucleo(s)tide analogue withdrawal depends on HBV genotype and viral antigen levels. J Hepatol. 2022;76:1042–1050. [DOI] [PubMed] [Google Scholar]
- 10. Seto WK, Hui AJ, Wong VW, Wong GL, Liu KS, Lai CL, et al. Treatment cessation of entecavir in Asian patients with hepatitis B e antigen negative chronic hepatitis B: a multicentre prospective study. Gut. 2015;64:667–672. [DOI] [PubMed] [Google Scholar]
- 11. Jeng WJ, Chen YC, Sheen IS, Lin CL, Hu TH, Chien RN, et al. Clinical relapse after cessation of tenofovir therapy in hepatitis B e antigen-negative patients. Clin Gastroenterol Hepatol. 2016;14:1813–1820 e1811. [DOI] [PubMed] [Google Scholar]
- 12. Wang CC, Tseng KC, Hsieh TY, Tseng TC, Lin HH, Kao JH. Assessing the durability of entecavir-treated hepatitis b using quantitative HBsAg. Am J Gastroenterol. 2016;111:1286–1294. [DOI] [PubMed] [Google Scholar]
- 13. Jeng WJ, Chen YC, Chien RN, Sheen IS, Liaw YF. Incidence and predictors of hepatitis B surface antigen seroclearance after cessation of nucleos(t)ide analogue therapy in hepatitis B e antigen-negative chronic hepatitis B. Hepatology. 2018;68:425–434. [DOI] [PubMed] [Google Scholar]
- 14. Berg T, Simon KG, Mauss S, Schott E, Heyne R, Klass DM, et al. Long-term response after stopping tenofovir disoproxil fumarate in non-cirrhotic HBeAg-negative patients - FINITE study. J Hepatol. 2017;67:918–924. [DOI] [PubMed] [Google Scholar]
- 15. Liem KS, Fung S, Wong DK, Yim C, Noureldin S, Chen J, et al. Limited sustained response after stopping nucleos(t)ide analogues in patients with chronic hepatitis B: results from a randomised controlled trial (Toronto STOP study). Gut. 2019;68:2206–2213. [DOI] [PubMed] [Google Scholar]
- 16. Liu J, Li T, Zhang L, Xu A. The role of hepatitis b surface antigen in nucleos(t)ide analogues cessation among asian patients with chronic hepatitis B: A systematic review. Hepatology. 2019;70:1045–1055. [DOI] [PubMed] [Google Scholar]
- 17. Hadziyannis SJ, Sevastianos V, Rapti I, Vassilopoulos D, Hadziyannis E. Sustained responses and loss of HBsAg in HBeAg-negative patients with chronic hepatitis B who stop long-term treatment with adefovir. Gastroenterology. 2012;143:629–636 e621. [DOI] [PubMed] [Google Scholar]
- 18. Jeng WJ, Hsu YC, Su TH, Chen CH. Letter to the Editor: Deliberations more than a cut-off hepatitis B surface antigen value for nucleos(t)ide analogue cessation. Hepatology. 2019;70:1488–1489. [DOI] [PubMed] [Google Scholar]
- 19. Scholtes C, Hamilton AT, Plissonnier ML, Charre C, Scott B, Wang L, et al. Performance of the cobas(R) HBV RNA automated investigational assay for the detection and quantification of circulating HBV RNA in chronic HBV patients. J Clin Virol. 2022;150-151:105150. [DOI] [PubMed] [Google Scholar]
- 20. Fan R, Zhou B, Xu M, Tan D, Niu J, Wang H, et al. Association between negative results from tests for HBV DNA and RNA and durability of response after discontinuation of nucles(t)ide analogue therapy. Clin Gastroenterol Hepatol. 2020;18:719–727.e717. [DOI] [PubMed] [Google Scholar]
- 21. Tsuge M, Murakami E, Imamura M, Abe H, Miki D, Hiraga N, et al. Serum HBV RNA and HBeAg are useful markers for the safe discontinuation of nucleotide analogue treatments in chronic hepatitis B patients. J Gastroenterol. 2013;48:1188–1204. [DOI] [PubMed] [Google Scholar]
- 22. van Bommel F, Bartens A, Mysickova A, Hofmann J, Kruger DH, Berg T, et al. Serum hepatitis B virus RNA levels as an early predictor of hepatitis B envelope antigen seroconversion during treatment with polymerase inhibitors. Hepatology. 2015;61:66–76. [DOI] [PubMed] [Google Scholar]
- 23. Kramvis A, Chang KM, Dandri M, Farci P, Glebe D, Hu J, et al. A roadmap for serum biomarkers for hepatitis B virus: current status and future outlook. Nat Rev Gastroenterol Hepatol. 2022;19:727–745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Jackson K, Bonanzinga S, Edwards R, Visvanathan K, Li X, Hall S, et al. Assessment of the cobas HBV RNA Investigational Assay in the Setting of Nucleoside Analogue Therapy Cessation. 2022. [DOI] [PubMed]
- 25. Cortese MF, Riveiro-barciela M, Tabernero D, Rodriguez-algarra F, Palom A, Sopena S, et al. standardized hepatitis B virus RNA quantification in untreated and treated chronic patients: A promising marker of infection follow-up. Microbiol Spectr. 2022;10:e0214921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Freeman GH, Halton JH. Note on an exact treatment of contingency, goodness of fit and other problems of significance. Biometrika. 1951;38:141–149. [PubMed] [Google Scholar]
- 27. Gray RJ. A class of K-sample tests for comparing the cumulative incidence of a competing risk. Ann Statist. 1988;16:1141–1154. [Google Scholar]
- 28. Pandey M, Jain A. ROC Curve: Making way for correct diagnosis. Pharma SUG 2016 May 8-11. Denver, Colorado. 2016. [Google Scholar]
- 29. Marcellin P, Wong DK, Sievert W, Buggisch P, Petersen J, Flisiak R, et al. Ten-year efficacy and safety of tenofovir disoproxil fumarate treatment for chronic hepatitis B virus infection. Liver Int. 2019;39:1868–1875. [DOI] [PubMed] [Google Scholar]
- 30. Ahn SH, Marcellin P, Ma X, Caruntu FA, Tak WY, Elkhashab M, et al. Hepatitis B surface antigen loss with tenofovir disoproxil fumarate plus peginterferon alfa-2a: Week 120 analysis. Dig Dis Sci. 2018;63:3487–3497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hirode G, Choi HSJ, Chen CH, Su TH, Seto WK, Van Hees S, et al. Off-therapy response after nucleos(t)ide analogue withdrawal in patients with chronic hepatitis B: An international, multicenter, multiethnic cohort (RETRACT-B Study). Gastroenterology. 2022;162:757–771 e754. [DOI] [PubMed] [Google Scholar]
- 32. Deng R, Liu S, Shen S, Guo H, Sun J, Circulating HBV. RNA: From biology to clinical applications. Hepatology. 2022;76:1520–1530. [DOI] [PubMed] [Google Scholar]
- 33. Liu S, Zhou B, Valdes JD, Sun J, Guo H. Serum hepatitis B virus RNA: A new potential biomarker for chronic hepatitis B virus infection. Hepatology. 2019;69:1816–1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Farag MS, van Campenhout MJH, Pfefferkorn M, Fischer J, Deichsel D, Boonstra A, et al. Hepatitis B virus RNA as early predictor for response to pegylated interferon alpha in HBeAg-negative chronic hepatitis B. Clin Infect Dis. 2021;72:202–211. [DOI] [PubMed] [Google Scholar]
- 35. van Campenhout MJH, van Bommel F, Pfefferkorn M, Fischer J, Deichsel D, Boonstra A, et al. Host and viral factors associated with serum hepatitis B virus RNA levels among patients in need for treatment. Hepatology. 2018;68:839–847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Laras A, Papatheodoridi M, Panopoulou E, Papatheodoridis GV, Hadziyannis SJ, Hadziyannis E. Serum hepatitis B virus RNA detectability, composition and clinical significance in patients with ab initio hepatitis B e antigen negative chronic hepatitis B. Virol J. 2022;19:22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Butler EK, Gersch J, McNamara A, Luk KC, Holzmayer V, de Medina M, et al. Hepatitis B virus serum DNA andRNA levels in nucleos(t)ide analog-treated or untreated patients during chronic and acute infection. Hepatology. 2018;68:2106–2117. [DOI] [PubMed] [Google Scholar]
- 38. Giersch K, Allweiss L, Volz T, Dandri M, Lutgehetmann M. Serum HBV pgRNA as a clinical marker for cccDNA activity. J Hepatol. 2017;66:460–462. [DOI] [PubMed] [Google Scholar]
- 39. Dandri M, Petersen J. cccDNA maintenance in chronic hepatitis B - targeting the matrix of viral replication. Infect Drug Resist. 2020;13:3873–3886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Belloni L, Pollicino T, De Nicola F, Guerrieri F, Raffa G, Fanciulli M, et al. Nuclear HBx binds the HBV minichromosome and modifies the epigenetic regulation of cccDNA function. Proc Natl Acad Sci U S A. 2009;106:19975–19979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Decorsiere A, Mueller H, van Breugel PC, Abdul F, Gerossier L, Beran RK, et al. Hepatitis B virus X protein identifies the Smc5/6 complex as a host restriction factor. Nature. 2016;531:386–389. [DOI] [PubMed] [Google Scholar]
- 42. Sekiba K, Otsuka M, Ohno M, Yamagami M, Kishikawa T, Suzuki T, et al. Inhibition of HBV transcription from cccDNA with nitazoxanide by targeting the HBx-DDB1 interaction. Cell Mol Gastroenterol Hepatol. 2019;7:297–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Mitra B, Guo H. Hepatitis B virus X protein crosses out Smc5/6 complex to maintain covalently closed circular DNA transcription. Hepatology. 2016;64:2246–2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Fu X, Song X, Li Y, Tan D, Liu G. Hepatitis B virus X protein upregulates DNA methyltransferase 3A/3B and enhances SOCS-1CpG island methylation. Mol Med Rep. 2016;13:301–308. [DOI] [PubMed] [Google Scholar]
- 45. Kim JW, Lee SH, Park YS, Hwang JH, Jeong SH, Kim N, et al. Replicative activity of hepatitis B virus is negatively associated with methylation of covalently closed circular DNA in advanced hepatitis B virus infection. Intervirology. 2011;54:316–325. [DOI] [PubMed] [Google Scholar]
- 46. Zhang Y, Mao R, Yan R, Cai D, Zhang Y, Zhu H, et al. Transcription of hepatitis B virus covalently closed circular DNA is regulated by CpG methylation during chronic infection. PLoS One. 2014;9:e110442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Hong X, Kim ES, Guo H. Epigenetic regulation of hepatitis B virus covalently closed circular DNA: Implications for epigenetic therapy against chronic hepatitis B. Hepatology. 2017;66:2066–2077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Dandri M. Epigenetic modulation in chronic hepatitis B virus infection. Semin Immunopathol. 2020;42:173–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Carey I, Gersch J, Wang B, Moigboi C, Kuhns M, Cloherty G, et al. Pregenomic HBV RNA and hepatitis B Core-related antigen predict outcomes in hepatitis B e antigen-negative chronic hepatitis B patients suppressed on nucleos(t)ide analogue therapy. Hepatology. 2020;72:42–57. [DOI] [PubMed] [Google Scholar]


