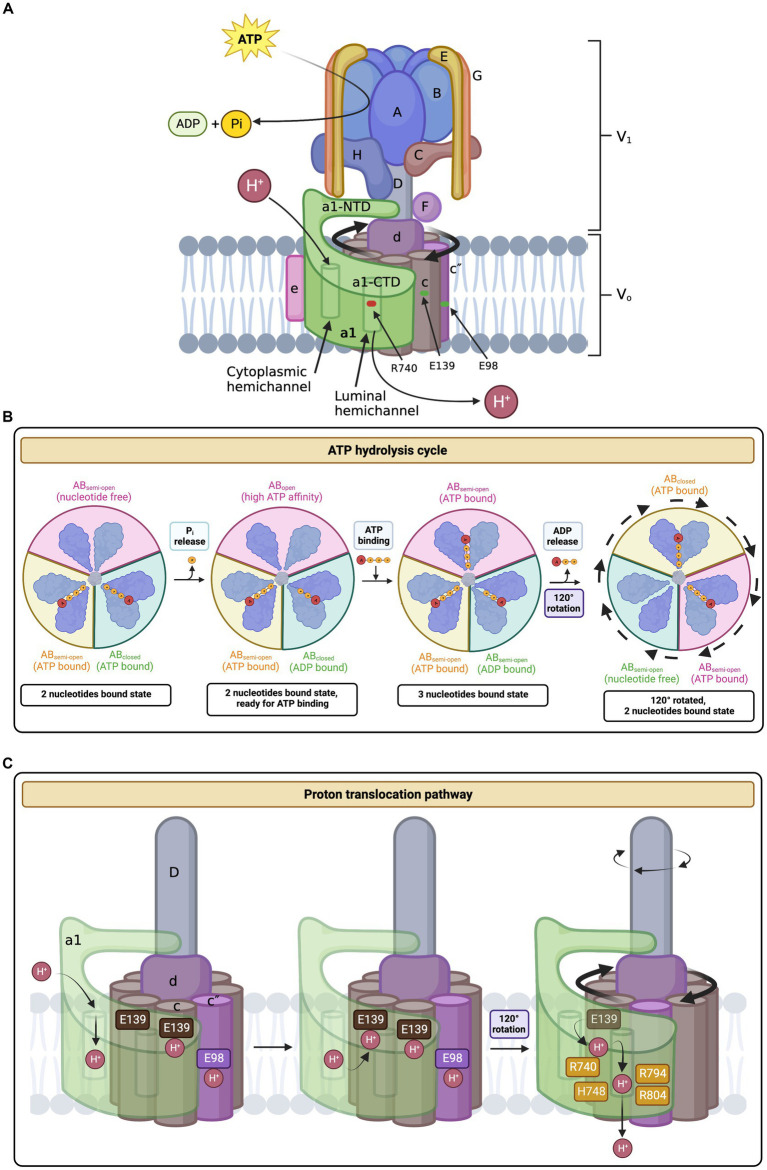
Figure 1.
Structure and mechanism of the human V-ATPase. (A) V-ATPase structure. The cytoplasmic V1 sector is responsible for ATP hydrolysis and contains the following subunits: A3, B3, C, D, E3, F, G3, and H. The membrane-embedded Vo sector mediates proton translocation and is composed of the following subunits: a1 (neuronal isoform), c9, c″, d, e, Ac45 (also known as ATP6AP1), RNaseK, ATP6AP2. The top of V1 includes the A3B3 hexamer head, the site of ATP binding and hydrolysis. ATP hydrolysis induces the rotation of the A3B3 hexamer which subsequently powers the rotation of the c-ring (composed of subunits c9 and c″) via linkage through subunits D, F, and d (the central stalk). The peripheral stalks, composed of subunits E and G, act as stators and connect the V1 sector with Vo. Protons enter and exit subunit a1 via the hemichannels and interact with key residues in a1 and the c-ring in the process of proton translocation. (B) ATP hydrolysis cycle. ATP hydrolysis occurs between the interface of the A and B subunits in the V1 complex. The C-terminal domains of A and B undergo conformational changes depending on their bound state to ATP and ADP. First, ATP is tightly bound between the A and B subunits and is subsequently hydrolyzed into ADP and inorganic phosphate (Pi). The Pi is released and the AB pair changes conformations to a less tight ADP-bound form. This triggers the adjacent AB pair to change conformations from a semi-open (nucleotide-free) conformation to a high ATP-affinity open confirmation. Next, ATP binds tightly to the open AB pair, changing the ADP-bound dwell into a semi-open conformation, thus releasing ADP. The conformational changes associated with the ATP hydrolysis cycle induce the tilting and oscillation of the A3B3 hexamer. (C) Proton translocation pathway. A proton enters the cytoplasmic hemichannel and protonates an essential glutamic acid residue on the c-ring (E139 subunit c or E98 subunit c”). Next, the c-ring, powered by ATP hydrolysis in V1, rotates clockwise through the lipid bilayer to bring the protonated c-ring glutamates close to an essential a1 residue, R740. A salt bridge forms between the protonated glutamate residue and R740, causing the release of the proton into the luminal hemichannel. Finally, the proton exits the membrane and enters the lumen by following a network of polar and negatively charged residues lining the luminal hemichannel (H748, E794, and R804), resulting in luminal acidification. For every three ATP molecules hydrolyzed, 10 protons are translocated. Amino acid positions are based on the updated accession number NM_001130021.3. The figure was created with BioRender.com.