Figure 3.
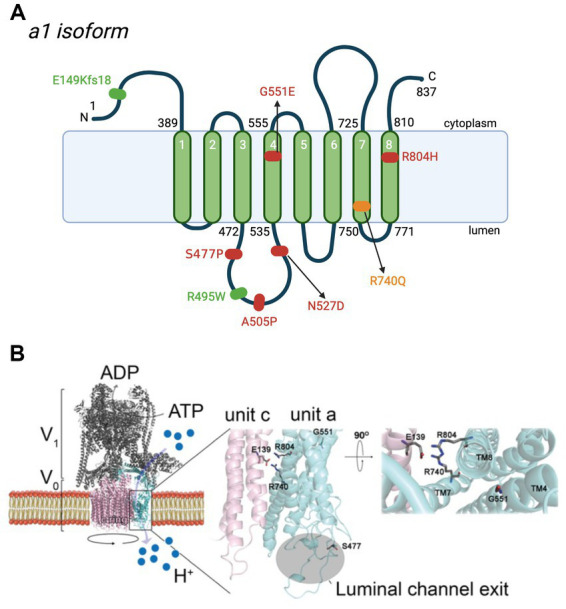
De novo and biallelic mutations to ATP6V0A1 impair V-ATPase functions in cases of DEE and PME. (A) Schematic representation of the location of mutations to subunit a1 in DEE (red), early onset PME (green), and both diseases (yellow) (Aoto et al., 2021; Bott et al., 2021). The most common mutation is R740Q, which is de novo, heterozygous, and follows a dominant inheritance pattern. Other de novo heterozygote variants are S477P, G551E, and R804H. Biallelic mutations, E149Kfs18 and R495W, are reported in 5 individuals with PME, 4 of whom are from the same family. In DEE individuals, A505P and N527D are in compound heterozygosity, each with a 50-kb deletion [del(17)(q21.2)] and a splice site mutation (c.196C + 1G > A), respectively. Positions are based on the updated accession number NM_001130021.3. For the purpose of annotation, transmembrane α7-α8 are not tilted. Amino acid sequences are based on Q93050 in UniProt. (A) was created with BioRender.com. (B) Homology model of human V-ATPase and locations of residues reported in de novo mutations on subunit a1 (Bott et al., 2021). Residues S477, G551, R740, and R804 are represented as stick structures relative to the key glutamic acid residue, E139 of subunit c. S477, at the second short linker loop (connecting transmembrane α3-α4) of luminal hemichannel.
