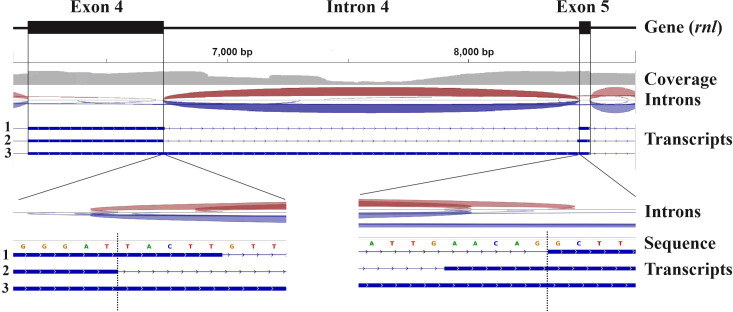
Figure 1.
Mapping of RNA-seq data to the mitochondrial genome of Coccidioides posadasii. The figure depicts a 2,579 nt window over the coordinates of exons 4 (6172-6736) and 5 (8458-8504) of the mitochondrial gene encoding the large subunit rRNA (rnl) of C. posadasii (see (de Melo Teixeira et al., 2021) for details on genome sequencing) as determined by rRNA sequence conservation and structural modelling of exons 4 and 5 plus adjacent introns. The read-mapping coverage distribution (grey) is shown below the annotated exons. Red and blue arches indicate forward and reverse split reads, respectively. The bottom part of the upper panel depicts the splice junctions inferred from the RNA-Seq read to genome mapping. The coverage profile reveals a substantial number of reads that map within intron 4 due to stable transcripts encoding intron ORFs and an elevated level of un-spliced RNA precursors, which is common in mitochondria. The two distinct splice junctions inferred at the 5’ and 3’ end of intron 4 are supported in roughly equal proportions by mapped reads. The bottom track depicts exons inferred from three distinct transcripts (marked 1, 2 and 3) that were reconstructed from the mapped reads using StringTie. The two lower panels show read mapping in higher resolution. The vertical dotted line indicates the predicted precise splice junction.