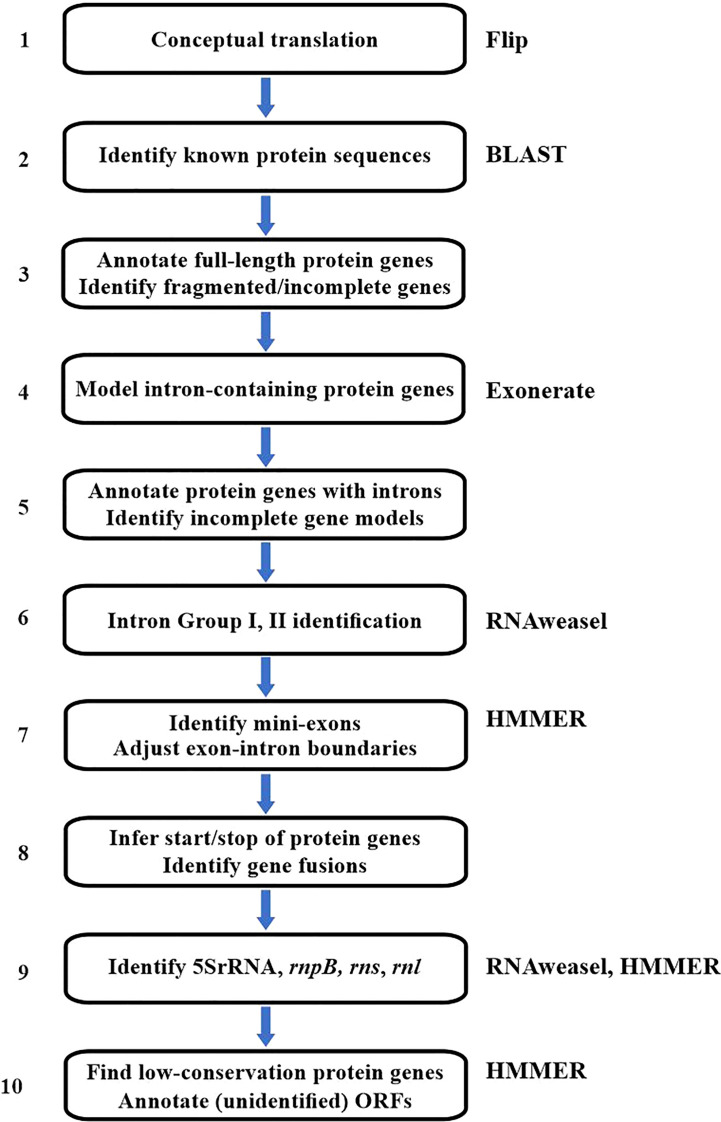
Figure 2.
Flowchart of the MFannot annotation procedure. The figure summarizes the analytical steps and the external tools employed (indicated to the right). The external tools are Flip (conceptual translation), BLAST (fast sequence similarity search), Exonerate (annotation of genes with exons/intron structure), HMMER (high sensitivity sequence matching), and RNAweasel (introns Group I and II, ncRNAs). When no specific tool is mentioned, the corresponding step is executed by MFannot-specific code. The RNAweasel tool (step 6 and 9) uses ERPIN (and more recently, Infernal) as a search engine. ERPIN’s search algorithm is based on RNA secondary structure profiles computed from RNA sequence alignments plus user-defined secondary-structure information as an input. Much of ERPIN’s efficiency stems from the definition of precisely delimited structural elements that can be searched individually or in combination using a particular search order (‘search strategy’). In general, ERPIN (Gautheret and Lambert, 2001) is the second-most sensitive search algorithm for structured RNAs [following the covariance-based Infernal program (Nawrocki and Eddy, 2013)]. Still, it distinguishes more reliably certain mitochondrial Group I and II introns.