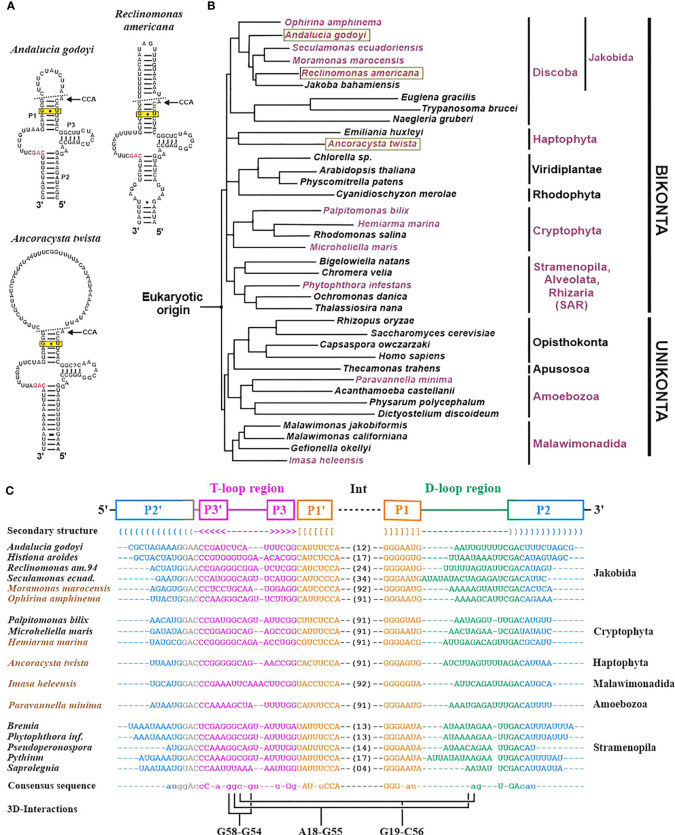
Figure 4.
Identification of mitochondrial tmRNA (ssrA) genes across eukaryotes. (A) Examples of mitochondrial tmRNA 2D structures. The helices P1, P2 and P3 are indicated in the A. godoyi 2D structure, in accordance with the sequence alignment in (C). The broken line in the 2D structure marks the sites of endonucleolytic processing that give rise to mature tRNA-like 5′ and 3′ ends. The 3′ end undergoes further modification by adding a CCA [51], as indicated in the figure. The invariant G-U pair in helix 3 (boxed) is required to recognize the tmRNA by tRNA synthase alanine. The GAC motif that is invariant across tmRNAs is marked in red. (B) Schematic eukaryotic tree (consensus derived from topologies in (Derelle and Lang, 2011; Derelle et al., 2015; Derelle et al., 2016; Janouškovec et al., 2017; Heiss et al., 2021; Tekle et al., 2022; Yazaki et al., 2022)). Names of species containing a mt ssrA gene are marked in mauve, and the three species with 2D structures shown in (A) are boxed. Mt ssrA genes were identified by an exhaustive search of published mitogenomes with a previously published covariance model based on genes from jakobids plus stramenopiles (Hafez et al., 2013a). The search identifies homologs in four eukaryotic lineages formerly thought to lack mt ssrA (cryptophytes, haptophytes, malawimonads, and amoebozoans), as well as in numerous additional Oomycota (represented in the tree by a single species, Phytophthora infestans). (C) Structured alignment of tmRNA sequences. Brown text colour marks the six species for which an ssrA annotation in GenBank records is lacking (Shiratori and Ishida, 2016; Strassert et al., 2016; Janouškovec et al., 2017; Yabuki et al., 2018; Bondarenko et al., 2019; Heiss et al., 2021). For further details on previous results and the structural annotation, see (Hafez et al., 2013a). Note that a new version of MFannot (available in June 2023) includes tmRNA-CM searches. During the preparation of the manuscript, we noted a recent publication on two additional jakobid mitogenomes that does not mention the presence of ssrA genes (Galindo et al., 2023). As expected due to its almost ubiquitous presence across jakobids (the only current exception is J. bahamiensis), ssrA genes are present in both Agogonia voluta and Ophirina chinija mtDNAs (between rps2 and trnS(gcu); E-values of 6.7e-11 and 4.4e-13, respectively).