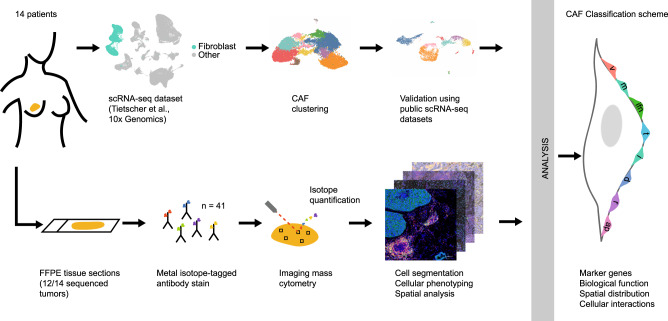
Fig. 1. Workflow used to define the CAF classification system.
scRNA-seq data from matched breast cancer samples25 were analysed for fibroblast heterogeneity with subsequent validation of identified fibroblast subtypes in other tumour types. IMC using formalin-fixed paraffin-embedded tissue sections (FFPE) was used to validate findings at the protein level and to evaluate spatial distribution. The resulting CAF classification system (featuring vascular CAFs (vCAFs), matrix CAFs (mCAFs), interferon-response CAFs (ifnCAFs), tumour-like CAFs (tCAFs), inflammatory CAFs (iCAFs), dividing CAFs (dCAFs), reticular-like CAFs (rCAFs) and antigen-presenting CAFs (apCAFs)) was based on marker genes, biological functions, spatial distribution within the TME, and cellular interactions.