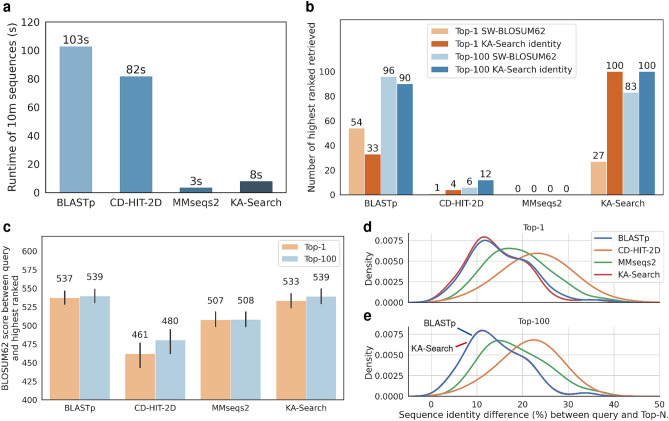
Figure 3.
Speed and sensitivity comparison between KA-Search and the commonly used protein search tools BLASTp (version 2.13.0)31, CD-Hit-2d (version 4.8.1)32 and MMseqs2 (version 13.45111)33. The default settings were used for each tool, and each search was done against the whole variable domain. BLOSUM62 scores were calculated by aligning with the Smith-Waterman algorithm using BLOSUM6234 as the substitution matrix and gap costs of 11 for existence and 1 for extension. (a) Runtime was calculated as the average time it took to search 10 million sequences (OAS-test) with a single query on a single CPU. Sensitivity was compared by the tools ability to find the exact closest match and highest similarity match. (b) Sensitivity was calculated by how often each tool returned the closest or the closest within the top-100 match for 100 heavy variable domains (test queries) against the same 10 million sequences. The closest match was defined using two different identity metrics, the highest BLOSUM62 score and KA-Search identity. (c) Highest similarity, was first compared with the average BLOSUM62 score between the test queries and target sequences from the top-1 and the closest within the top-100 returned sequences from each search tool. Highest similarity was then compared using the density of sequence identity difference between the test queries and, (d) the top-1 returned sequences and, (e) the closest within the top-100 returned sequences from each search tool. For top-100, the density of BLASTp and KA-Search are nearly perfectly overlapping. Sequence identity was calculated as the percentage of exact matches after alignment with the Smith-Waterman algorithm using BLOSUM62 as the substitution matrix and gap costs of 11 for existence and 1 for extension.