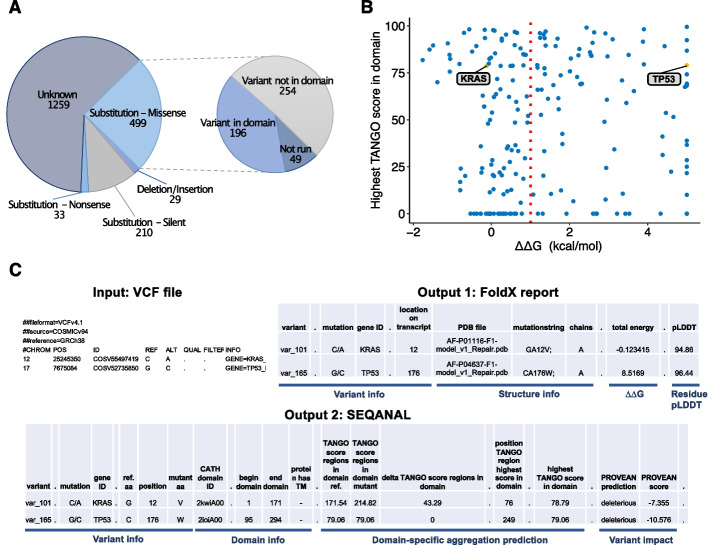
Fig. 3.
SNPeffect analysis of carcinoma cell line SHP-77. A Number of different mutation types in the small cell lung carcinoma cell line SHP-77. The impact of 90% of all missense variants was defined with SNPeffect 5.0. B Variant impact plot for the small cell lung carcinoma cell line SHP-77. The APR TANGO score and FoldX scores (maxed at ΔΔG = 5 in the plot) are plotted for all variants in a high-confidence domain. The two variants in known driver proteins, KRAS and p53, are highlighted in the plot. For variants that were matched to more than one PDB structure, only one structure (highest pLDDT) was used to avoid redundancy. C Condensed input and output files of cell line SHP-77 focusing on KRAS and p53. The FoldX report contains the calculated impact on structural stability (ΔΔG), information about the used structure, and the residue pLDDT score. The SEQANAL report presents the variant-containing domain, identification of APRs in that domain, and variant-impact prediction by established tools such as PROVEAN. Dotted columns represent extra data that can be found in the complete output files (Additional file 2)