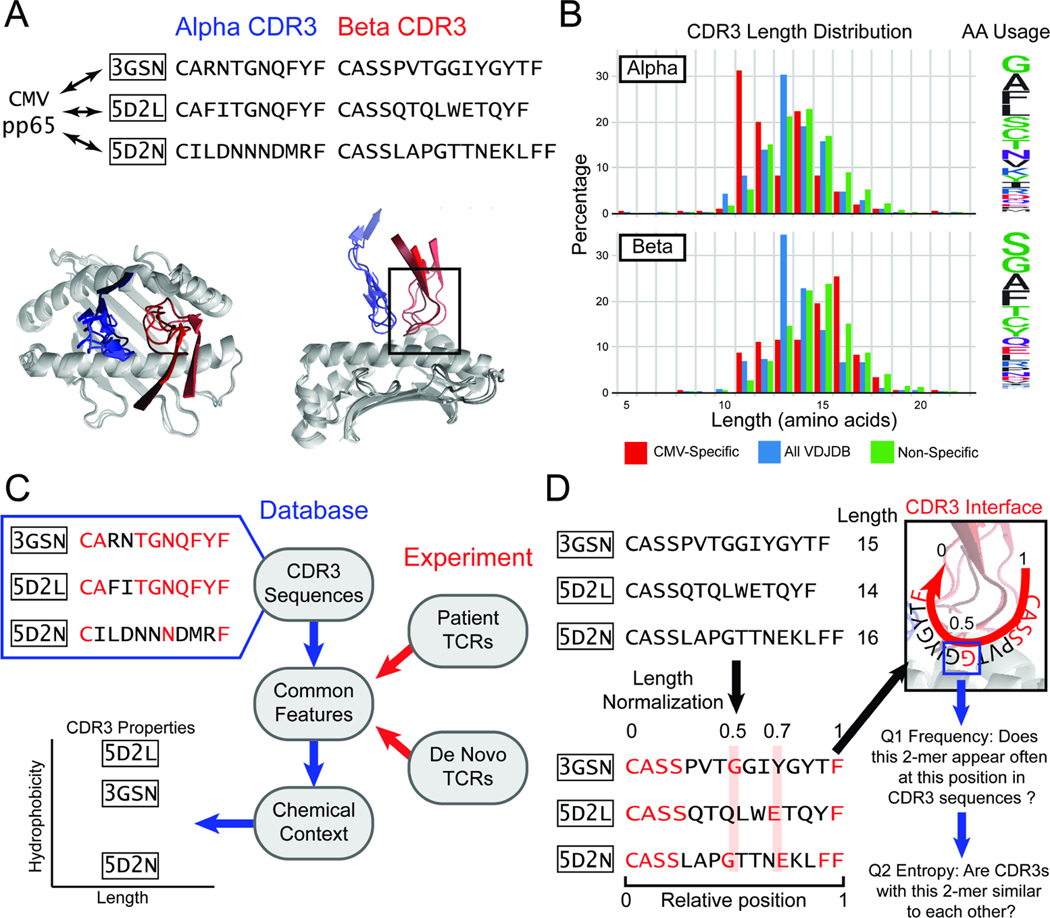
Figure 1. A length-agnostic framework to characterize TCRs.
A. Crystal structures of different CMV pp65-specific TCRs show that TCR (blue) and (red) chains bind in similar conformations. B. A wide range of CDR3 lengths are reported for sets of TCRs that bind to a specific target (red, CMV pp65 epitope), TCRs reported in the VDJdb database (blue), and sequenced TCRs from a human donor (green). The relative frequency of amino acids across the entire CDR3 is shown on the Logo plot (right). C. CDR3 sequences that bind the same target share common amino acid features (highlighted in red). The chemical contexts of these features such as CDR3 length or amino acid hydrophobiciity are associated with specificity to antigen. D. TCRs of different lengths are difficult to compare. SPAN-TCR normalizes the length of TCR CDR3s, then searches for amino acids or k-mer subsequences at similar positions, which may be likely to interact with the same section of the p-MHC (highlighted in red). For each k-mer, we determine first if the k-mer appears frequently at a position. Next, we determine if sequences that have the k-mer at the position are diverse or repetitive using entropic analysis.