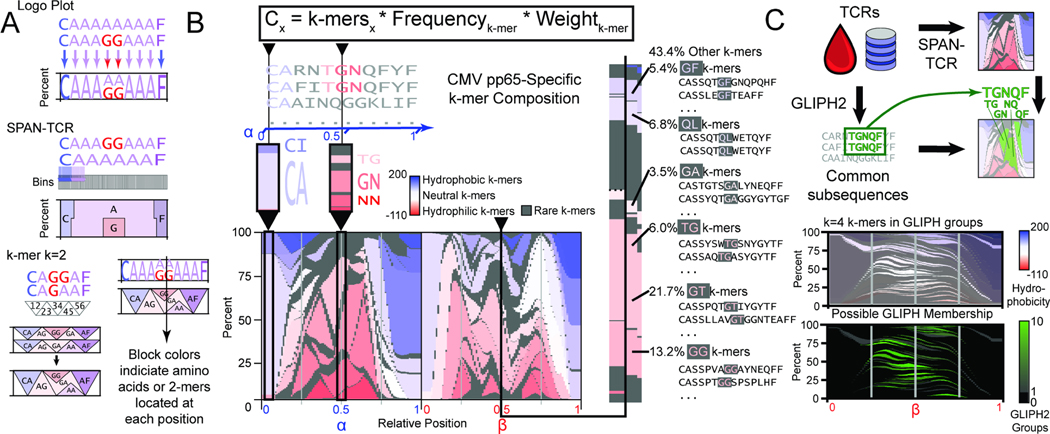
Figure 2.
The landscape of amino acid usage within CMV pp65 antigen-specific TCRs. A. SPAN-TCR analyzes the amino acid composition of TCR sets using bins. By increasing bin number from 10 to 100, differences are finely resolved. SPAN-TCR further targets k consecutive amino acids (k-mers). The weighted influence of a k-mer is strongest at its center and decays as the next consecutive k-mer increases its influence. B. The SPAN-TCR formula used to generate a weighted display of k-mers for k=2. Low frequency (<3%) 2-mers are shown in gray and otherwise colored by hydrophobicity (linear scale, hydrophobicity index relative to glycine). Common 2-mers near the center of the chain, and the CDR3s containing those 2-mers are shown. C. Comparison of SPAN-TCR and GLIPH. GLIPH identifies enriched subsequences, which can be projected onto SPAN-TCR representations of TCR sets. SPAN-TCR k-mers for k=4 found in GLIPH groups, and number of GLIPH groups each 4-mer belongs to, are plotted.