Figure 4.
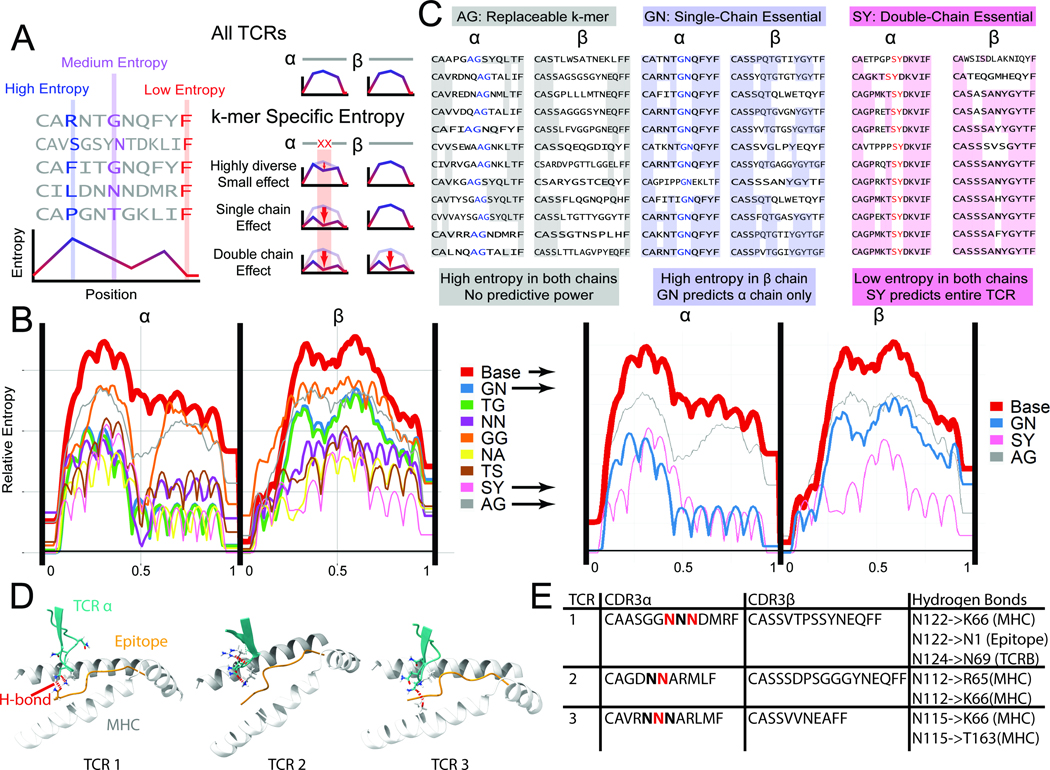
Entropy-based analysis of TCR repertoires. A. In a set of TCR sequences, the entropy at each position measures the diversity of amino acid (or 2-mer) usage. Low entropy at a position indicates repeated usage of 2-mers, possibly important for epitope binding and recognition. To determine if a 2-mer is important for binding, the subset of TCRs containing a 2-mer is isolated. Entropy calculated at every position can be unaffected in the subset relative to the whole set, reduced in the same chain, or reduced in both chains. B. The entropy for all CMV pp65-specific TCRs, as well as subsets of antigen-specific TCRs with common 2-mers near the center of the chain are plotted (N>10 TCRs per 2-mer). We observe 2-mers for which the set of CDR3s containing that 2-mer at a specific location has very low entropy (i.e. SY, NA, NN), denoting a specific sequence used for binding. The thickness of the lines is correlated to the number of TCRs containing the 2-mer at the center of the chain. C. 2-mers with minimal entropy reduction (AG), single-chain reduction (GN, essential), and double-chain reduction (SY, super-essential) are shown. See also Figs. S3–5. D. Simulated TCR-pMHC complexes are shown at the -chain CDR3/epitope/MHC interface. For the essential 2-mer NN, hydrogen bonds (red dashed lines) were found between the N residues and the MHC, the epitope, and/or the TCR chain. E. A tabulation of the specific H-bonding interactions observed in the three structures of panel D.
