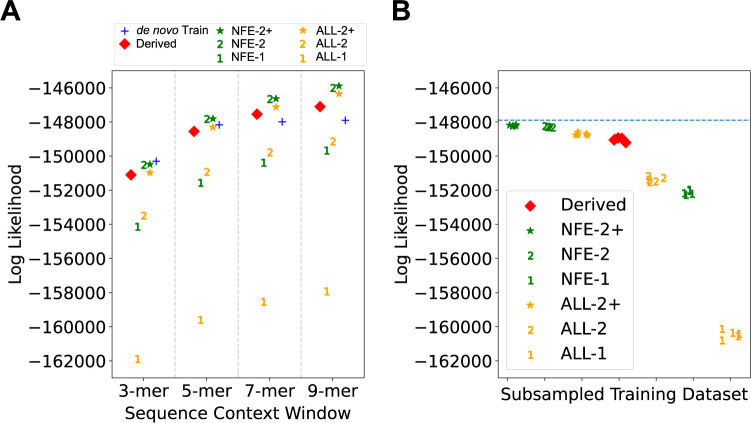
Fig 4. Modeling de novo mutation probabilities using polymorphism datasets.
Even base pair Halldorsson et al. de novo training data modeled by Baymer is compared to Baymer-modelled polymorphism datasets partitioned by allele count. (A) Multinomial likelihoods for each model are calculated on odd de novo base pair test data at various sequence context sizes. Polymorphism probability estimates were linearly scaled to match the mean polymorphism probability of the holdout dataset. (B) Polymorphism datasets were down-sampled to match the size of the even base pair de novo data (70,364 variants) and multinomial likelihoods were calculated on odd de novo base pair data. Each dataset was down-sampled using 5 different random seeds. The Log Likelihood of the 9-mer de novo training model is indicated with the blue dotted line.