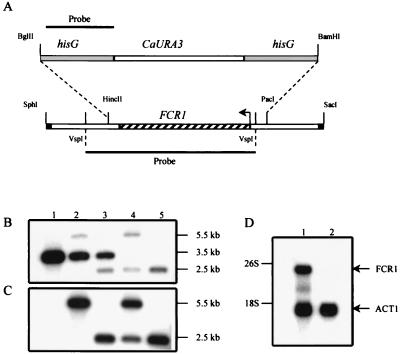
FIG. 6.
Chromosomal deletion of the FCR1 locus. (A) Schematic representation of the strategy used to generate the FCR1 deletion in the strain CAI4. The hisG-CaURA3-hisG cassette was used to replace the HincII-PacI fragment containing the FCR1 ORF. Only the relevant restriction sites are shown (the SphI and SacI sites are derived from the pGEM7Z-f(+) vector). The 0.9-kb hisG fragment (top) and the 2.3-kb VspI fragment (bottom) were used as probes to monitor the recombination events. (B and C) Southern blot analyses of genomic DNA from the parental CAI4 strain (lanes 1), the FCR1/fcr1Δ::hisG-CaURA3-hisG heterozygous strain (lanes 2), the FCR1/fcr1Δ::hisG strain after 5-FOA counter-selection (lanes 3), the fcr1Δ::hisG/fcr1Δ::hisG-CaURA3-hisG strain (lanes 4), and the fcr1Δ::hisG/fcr1Δ::hisG homozygous FM7 strain after the second 5-FOA counter-selection (lanes 5). The genomic DNA was digested with HindIII, electrophoresed in duplicate on an agarose gel, and transferred to nylon membranes. The blots were probed with the 2.3-kb VspI FCR1 fragment (B) or the 0.9-kb hisG fragment (C). Autoradiographies were for 10 and 15 h, respectively, with two intensifying screens at −80°C. The sizes of the fragments are indicated in kilobases. (D) Northern blot analysis of FCR1 in strains CAI4 and FM7. Total RNA (20 μg) prepared from the CAI4 (lane 1) and FM7 (lane 2) strains was electrophoresed on a 1% agarose gel, transferred to a nylon membrane, and probed simultaneously with an FCR1 probe and an ACT1 probe. Autoradiography was for 48 h, with two intensifying screens at −80°C.