FIG. 3.
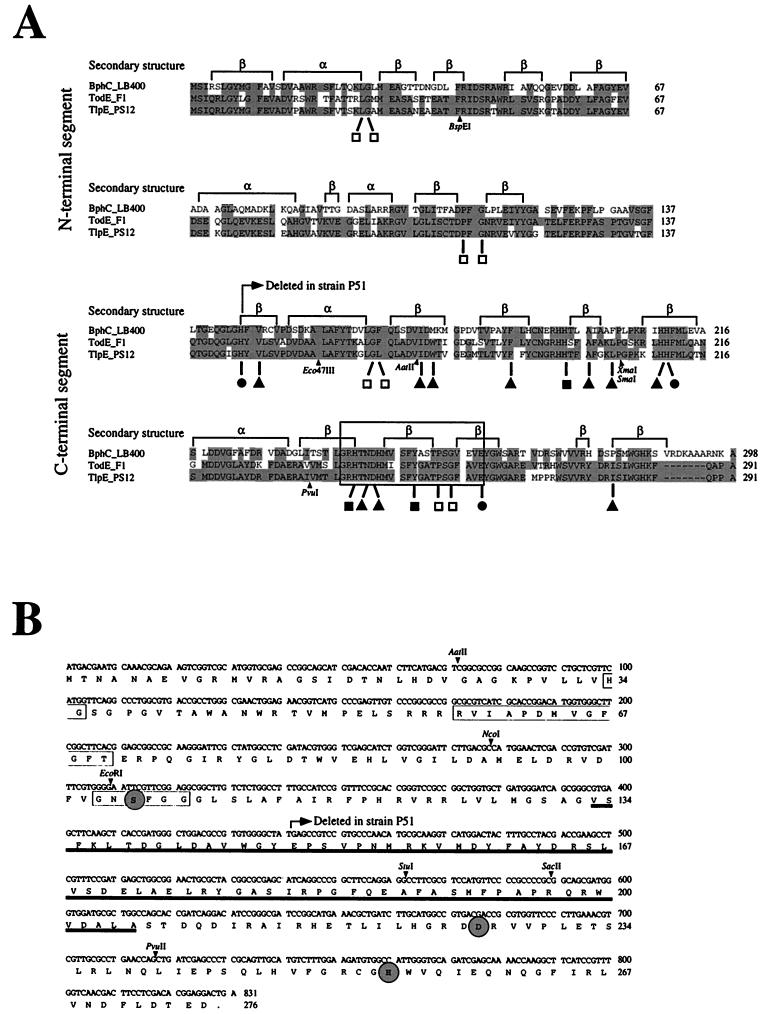
Sequence alignment and structural features of catechol 2,3-dioxygenases and nucleotide and deduced amino acid sequences of the TlpF hydrolase. (A) Alignment of extradiol dioxygenases of Burkholderia sp. strain LB400 (BphC_LB400) and P. putida F1 (TodE_F1) and the refunctionalized TlpE protein of Burkholderia sp. strain PS12 (TlpE_PS12). Based on the solved structure of the BphC enzyme, two domains (N and C terminal) with similar secondary structures can be distinguished (19), as indicated above the alignment. Identical residues are shaded. The amino acid ligands of the catalytic Fe(II) are marked (•), and those playing a direct catalytic role in the LB400 enzyme are indicated (■). Additional residues that form the substrate binding site in the LB400 enzyme are marked (▴), and conserved residues that play a structural role are indicated (□). The boxed fingerprint region contains the consensus sequence (G or N or T or I or V)-X1-H-X5 or 7-(L or I or V or M or F)-Y-X2-(D or E or N or T or A)-P-X1-(G or P)-X3 or 4-E (14), where Xn indicates n residues of any type, parentheses enclose residues found at one position, and boldface letters indicate the residues found in the TlpE protein. The positions of relevant restriction sites of the corresponding tlpE gene are indicated. (B) The dipeptide His-Gly of the putative oxanion hole, the RVIAPDXXGXGXS motif, and the so-called hydrolase or lipase box with the nucleophile motif Gly103-Xaa-Ser105-Xaa-Xaa-Gly108 (3, 11, 12) of the TlpF hydrolase are boxed. The catalytic residues Ser105, Asp226, and His254, representing the catalytic triad of hydrolases (2, 12, 29) and lipases (8, 11), are circled and in boldface. The region proposed to be involved in determination of substrate specificity (12) is underlined. Relevant restriction sites are indicated.