Summary
ERI1 is a 3′-to-5′ exoribonuclease involved in RNA metabolic pathways including 5.8S rRNA processing and turnover of histone mRNAs. Its biological and medical significance remain unclear. Here, we uncover a phenotypic dichotomy associated with bi-allelic ERI1 variants by reporting eight affected individuals from seven unrelated families. A severe spondyloepimetaphyseal dysplasia (SEMD) was identified in five affected individuals with missense variants but not in those with bi-allelic null variants, who showed mild intellectual disability and digital anomalies. The ERI1 missense variants cause a loss of the exoribonuclease activity, leading to defective trimming of the 5.8S rRNA 3′ end and a decreased degradation of replication-dependent histone mRNAs. Affected-individual-derived induced pluripotent stem cells (iPSCs) showed impaired in vitro chondrogenesis with downregulation of genes regulating skeletal patterning. Our study establishes an entity previously unreported in OMIM and provides a model showing a more severe effect of missense alleles than null alleles within recessive genotypes, suggesting a key role of ERI1-mediated RNA metabolism in human skeletal patterning and chondrogenesis.
Keywords: ERI1, skeletal dysplasia, ribosomopathy, spondyloepimetaphyseal dysplasia, short stature, exoribonuclease
Our study uncovers a ribosomopathy-related skeletal dysplasia caused by missense mutations in ERI1, which encodes an exoribonuclease. The findings set a precedent showing a more severe effect of missense alleles than null alleles within recessive genetic diseases and imply that ERI1-mediated RNA metabolism regulates development and homeostasis of human cartilage.
Introduction
ERI1 (MIM: 608739) encodes a 3′-to-5′ exoribonuclease that consists of an amino-terminal SAP domain (nucleic-acid-binding domain) and a carboxy-terminal 3′ exonuclease domain and belongs to the DEDDh family that includes poly(A) ribonuclease, RNase T, and the ϵ proofreading subunit of Escherichia coli DNA polymerase III.1 Eri1 is recruited into several conserved and species-specific RNA metabolic pathways. In Caenorhabditis elegans, ERI-1 forms a complex with Dicer that generates specific classes of endogenous siRNAs.2,3,4 In Schizosaccharomyces pombe, Eri1 regulates the turnover of chromatin-associated siRNAs.5 In the mouse, Eri1 targets three types of RNA molecules: rRNA, histone mRNA, and miRNA. Eri1 deficiency in mice results in abnormal 3′ end trimming of the 5.8S rRNA, impaired degradation of replication-dependent histone mRNAs, and increased abundance of mature miRNAs.6,7,8 The rRNA trimming function of ERI1 is conserved in Caenorhabditis elegans and Schizosaccharomyces pombe.9 However, the impact of ERI1 on these RNA molecules in humans remains unclear.
A homozygous 284 kb genomic deletion, which covers three genes (ERI1, MFHAS1, and MIR4660), was found in an individual with intellectual disability, cardiac malformation, and digit abnormalities.10 Recently, a homozygous nonsense variant in ERI1 was reported in an individual with developmental delay and distal limb abnormalities.11 This evidence supports the involvement of ERI1 in skeletal patterning and implies that a disorder is caused by the disruption of the exoribonuclease function.
In this study, we uncovered a phenotypic dichotomy in eight individuals from seven unrelated families with different types of ERI1 variants and highlighted the association of missense variants with spondyloepimetaphyseal dysplasia (SEMD), which is a group of skeletal diseases characterized by anomalies in spine and long tubular bones. In contrast, the affected individuals with bi-allelic null variants showed mild intellectual disability and digital anomalies. The detailed evaluation of the skeletal phenotypes of Eri1 knockout (KO) mice and the in vitro chondrogenesis of affected-individual-derived induced pluripotent stem cells (iPSCs) supported the functional involvement of ERI1 in skeletal development. Analyses using ERI1 KO HeLa cells and affected-individual-derived cells demonstrated functional conservation of ERI1 with its mouse ortholog in 5.8S rRNA maturation and histone mRNAs decay. The 5.8S rRNA maturation is involved in ribosome biogenesis, defects of which are known to cause ribosomopathies characterized by skeletal dysplasia.12,13,14,15 Our study leads to the findings of an SEMD associated with ribosomopathy and established a framework for understanding the molecular mechanisms underlying the ERI1 phenotypic dichotomy.
Material and methods
Next-generation sequencing
The study was approved by the ethical committee of RIKEN (approval number: 17-16-40(6) and data were shared in accordance to those of participating institutions. Genomic DNAs were extracted by standard procedures from peripheral blood of the individuals and their parents after informed consent. Exome sequencing (ES) and genome sequencing were performed as previously described.16,17,18,19,20,21,22 We searched for rare coding or splicing variants in disease-causing genes with transmissions consistent with Mendelian patterns of inheritance.
PCR and Sanger sequencing
Genomic fragments containing the variants identified by whole-exome sequencing (WES) were amplified by PCR and Sanger sequenced for both strands. The PCR primer sets are shown in Table S1.
RNA gel electrophoresis
Total RNAs were extracted from HeLa cells or the affected-individual-derived lymphoblastoid cells via SV Total RNA Isolation System (Promega). Small RNAs were extracted from HeLa cells via mirVana miRNA Isolation Kit (Thermo Fisher Scientific). 2.5 μg total RNAs or 3 μg small RNAs in Novex TBE-Urea Sample buffer (Thermo Fisher Scientific), Century Plus RNA Marker (Thermo Fisher Scientific), and microRNA Marker (New England Biolabs) were separated on 6% or 15% Novex TBE-Urea Gels in the Novex Mini-Cell (Thermo Fisher Scientific) for 60–70 min at a constant voltage of 180 V. Post-electrophoresis gels were stained by 2 μg/mL ethidium bromide for 20 min or SYBR Gold Nucleic Acid Gel Stain (Thermo Fisher Scientific) for 10 min in DEPC-treated water and images were captured under UV light.
Construction of expression plasmids
Clones for the coding sequences of ERI1 (GenBank: NM_153332.4) were PCR-amplified from a cDNA library derived from human peripheral blood via KOD -Plus- (Toyobo). The PCR amplicons were cloned into the Sal1 and NotI sites of the pCMV-HA expression vector (Clontech). The mutations were generated by inverse PCR-based site-directed mutagenesis kit (Toyobo). All PCR primers are shown in Table S1. HeLa cells were transfected with TransIT-LT1 (Mirus Bio) according the manufacturer’s instructions.
Generation of ERI1 KO HeLa cell line
Guide RNA (gRNA) sequences targeting genomic DNA of ERI1 were designed by E-CRISP (Figure S2A). Complementary oligos containing the gRNA sequences and BbsI ligation adapters were annealed and ligated into the BbsI-digested pX330 vector (Addgene #42230).23 The oligo sequences are listed in Table S1. We co-transfected the pair of pX330 plasmids with pEGFP-N1 (Clontech) vector, which we used as a fluorescent marker to sort transfected cells by FACSAria (BD Biosciences) (Figure S2B). We obtained monoclonal cell populations by limiting dilution of the sorted cells in half 96-well plate as performed previously.19 The cell lines with deletions covering exon 2 of ERI1 were identified by PCR for genomic DNA (Figure S2C) and confirmed by Sanger sequencing (Figure S2D). We performed western blot and RNA electrophoresis to validate the knockout effect (Figures S2E and S2F).
Western blot
Whole-cell lysates were harvested with 200 μL of RIPA Lysis and Extraction Buffer (Thermo Fisher Scientific). Western blots were performed by standard procedures with antibodies for ERI1 (1:100 dilution, sc-137089, Santa Cruz), β-actin-HRP (1:10,000 dilution, PM053-7, MBL, Japan), mouse IgG–peroxidase (1:10,000 dilution, #7076, Cell Signaling Technology), and HA-tag-HRP (1:1,000 dilution, #14031, Cell Signaling Technology). The signals were detected by ECL Prime Western Blotting Detection Reagents (RPN2232, GE Healthcare).
Real-time quantitative PCR
Affected-individual-derived fibroblasts and HeLa cells were treated with 5 mM hydroxyurea for 45 min. Total RNA from affected-individual-derived iPSCs and fibroblasts, and HeLa cells, was extracted with the SV Total RNA Isolation System (Promega) or QIAzol (QIAGEN) according to the manufacturer’s instructions. qPCR was performed as previously described.19,24 The qPCR primers are listed in Table S1.
Mice
Mice were housed in specific pathogen-free barrier facilities and used in accordance with protocols approved by the animal care and use committees of the Helmholtz Center, Munich and University of California San Francisco. We created the Eri1-deficient mice by gene targeting in BRUCE4 C57BL/6 ES cells by using standard techniques.7 Wild-type (WT) C57BL/6/J and Actin-FLPe transgenic mice (B6; SJLTg(ACTFLPe)9205Dym/J) were obtained from Jackson Laboratories, CMVCre transgenic mice (6.C-Tg(CMV-cre)1Cgn) were provided by K. Rajewsky (Harvard Medical School, Boston). To rescue mice from death, which affects offspring on a pure C57BL/6 background,7 we intercrossed mice once with the NMRI outbred strain background to then generate homozygous F2 mice. The mice used for blood cell tests were on a mixed ICR/C57BL/6 background, which were similarly generated by crossing ICR mice to Eri1+/− C57BL/6 mice and backcrossing F1 mice to Eri1+/− C57BL/6.6 Blood was collected in EDTA-coated tubes for red blood cell counts analyzed with a Hemavet system counter (CDC Technologies).
X-ray examination
Screening of bone phenotypes of the mice was performed at the age of 16 weeks. At the German Mouse Clinic (GMC), mice were maintained in individually ventilated cages with water and standard mouse chow according to German laws and GMC housing conditions.25 The skeletal morphology of the adult mice was analyzed with Faxitron X-ray Model MX-20 (Specimen Radiography System) equipped with an NTB Digital X-ray Scanner EZ 40 (NTBGmbH). Image processing was performed with ImageJ software.
Generation of iPSCs
Experimental use of affected-individual-derived cells was approved by ethics committees of RIKEN IMS (Yokohama). Informed consent was obtained from legal guardians of individuals by relevant institutions. The Epstein-Barr virus (EBV)-immortalized lymphoblastoid cell lines (LCLs) were generated from the peripheral blood lymphocytes of the individual and the parents as described previously.26 LCLs were cultured in Roswell Park Memorial Institute (RPMI) 1640 medium (Sigma) supplemented with 10% fetal bovine serum (Gibco Life Technologies). Reprogramming of LCLs was performed with episomes as previously described with a minor modification.27 Briefly, LCLs were electroporated with four episomal vectors (pCXLE-hOCT3/4-shp53-F, pCXLE-hSK, pCXLE-hUL, and pCXWB-EBNA1), which were gifts from Shinya Yamanaka (Addgene #27077, #27078, #27080, and #37624).28 The nucleofected LCLs were seeded on iMatrix-511 (Nippi)-coated plates containing TeSR-E7 reprogramming media (STEMCELL Technology) without feeder cells. After 18–21 days, iPSC colonies were manually picked and expanded in mTeSR-1 maintenance media (STEMCELL Technologies).27 The iPSCs were routinely passaged every 5–7 days at a ratio of 1:6 with TrypLE (Gibco Life Technologies).
Validation of the established iPSCs
Morphological images of iPSC colonies were captured with an Olympus CKK53 microscope with an UPlanFL N 4×/0.13na objective lens (Olympus) and Canon digital camera EOS Kiss X10 (Figure S3A). Expression of OCT3/4, SOX2, and NANOG mRNAs was confirmed by real-time quantitative PCR (Figure S3B). Primers sequences are provided in Table S1. Chromosomal G-banding analyses were performed by Chromocentre (Figure S3C). Established iPSCs were differentiated into ectoderm, mesoderm, and endoderm lineages via Pluripotent Stem Cell Functional Identification Kit (R&D Systems) according to the manufacturer’s protocol (Figure S3D). In brief, cells were fixed in 4% paraformaldehyde for 20 min at room temperature (RT), washed with PBS, and blocked for 45 min at RT with blocking solution (PBS + 1% BSA + 0.3% Triton-X-100 + 10% normal donkey serum). After blocking, cells were stained with primary antibodies overnight at 4°C. The next day, the cells were washed with PBS and incubated with Anti-Goat Alexa Fluor 488 secondary antibody (ab150129, Abcam) for 1 h at RT. Nuclei were stained with DAPI and the slides were mounted with fluorescent mounting medium (Dako). Images were acquired with a Nikon A1 microscope.
Chondrogenic differentiation of iPSCs
iPSCs were induced into chondrocytes as described previously.29 In brief, the iPSCs were initially differentiated into mesendodermal cells in DMEM/F12 (Gibco Life Technologies) with 10 ng/mL of Wnt-3a (R&D Systems), 10 ng/mL of Activin A (PeproTech), 1% ITS (Gibco Life Technologies), 1% FBS, and 50 units and 50 μg/mL of penicillin and streptomycin. After 3 days, the medium was changed to a chondrogenic medium that consists of DMEM with 1% ITS, 1% FBS, 2 mM L-glutamine, 1 × 10−4 M nonessential amino acids (Wako), 1 mM Na pyruvate (Sigma), 50 μg/mL of ascorbic acid (Wako), 10 ng/mL of BMP2 (Proteintech), 10 ng/mL of TGF-β1 (Proteintech), and 10 ng/mL of GDF5 (PeproTech), 50 units of penicillin, and 50 μg/mL of streptomycin. We added 10 ng/mL of bFGF (Wako) to the chondrogenic medium from day 3 to day 14 to increase the cell proliferation. The medium was changed every 2 days.
RNA sequencing (RNA-seq)
Total RNA from was extracted with the SV Total RNA Isolation System (Promega) according to the manufacturer’s instructions. Total RNAs were quantified and qualified by Qubit RNA Assay (Thermo Fisher) and TapeStation RNA ScreenTape/BioAnalyzer RNA Pico Kit (Agilent). RNA-seq library preparation was conducted with MGIEasy RNA Directional Library Prep Set V2.0 (MGI Tech). The libraries were fed into a DNBSEQ-G400 sequencer (MGI Tech) according to the manufacturer’s instructions, with paired end reads and mean read lengths of 150 bp. Read quality was assessed and adapters were trimmed from the reads via fastp v.0.20.0. RNA-seq yielded about 40 million reads per sample.
Differential gene expression analysis of RNA-seq data
The reads from RNA-seq were aligned to the human genome (GRCh38) with HISAT2 v 2.2.0. Aligned reads were quantified with featureCounts 2.6.0. Transcripts per million (TPM) counts was calculated by Python package bioinfokit v2.0.8. The volcano plot and the ridgeline plot were created with ExpressAnalyst. The heatmaps were created by GraphPad Prism 8.0. Enrichment analysis was performed by ExpressAnalyst.
Alcian blue staining
Cells were fixed with 95% methanol for 20 min and stained with 1% Alcian blue 8GX (Sigma) in 0.1 M HCL overnight.30 We washed the stained cells aggregated in plates at least three times with deionized water to guarantee only specific staining was left and we scanned these cells with a light microscope (Olympus CK40).
Statistical analysis
Continuous variables were reported as mean ± SD and compared with a two-tailed Student’s t test or a one-way ANOVA followed by a post-hoc Tukey’s multiple-comparison test as specified in the figure legends. p values < 0.05 were considered to be significant.
Results
A phenotypic dichotomy is identified in individuals with ERI1 variants
ERI1 variants were identified as candidates for the severe skeletal dysplasia by ES analysis independently for families 1, 2, and 3. It was identified as a candidate for the distal limb anomalies independently by ES analysis for families 5 and 6. Investigators were in contact through GeneMatcher. Family 4 was identified by a search of the Baylor Genetics Laboratories exome variant database for individuals with two ERI1 potentially deleterious variants, and family 7 was identified by a search of the 100,000 Genomes Project variant database. Six missense variants, three nonsense variants, and one splice-site variant of ERI1 were identified in these affected individuals (Table 1). Homozygosity or compound heterozygosity of these variants was confirmed in the individuals except individual 4, who harbored p.Ser21∗ and p.Pro155Leu, as the parents were not available. The frequency of these variants was less than 0.0001 (c.464C>T [p.Pro155Leu], c.893A>G [p.Asp298Gly]) or not present (the others) in gnomAD and ExAC.
Table 1.
Clinical and radiographic findings of the families with ERI1 variants
|
Individual with at least one missense variant |
Individual with bi-allelic null variant |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
|
Individual |
Individual 1A |
Individual 1B |
Individual 2 |
Individual 3 |
Individual 4 |
Individual 5 |
Individual 6 |
Individual 7 |
Hoxha et al.11 | Choucair et al.10 |
| Family | Family 1 | Family 1 | Family 2 | Family 3 | Family 4 | Family 5 | Family 6 | Family 7 | ||
| Varianta | ||||||||||
| DNA | c.[450A>T]; [893A>G] | c.[450A>T]; [893A>G] | c.[464C>T]; [893A>C] | c.[401A>G]; [895T>C] | c.[464C>T]; [62C>A] | c.[514C>T]; [514C>T] | c.[730C>T]; [730C>T] | c.[582+1G>A]; [582+1G>A] | c.[ 352A>T]; [352A>T] | g.[8783887_9068578del]; [8783887_9068578del] |
| Protein | p.[Glu150Asp]; [Asp298Gly] | p.[Glu150Asp]; [Asp298Gly] | p.[Pro155Leu]; [Asp298Ala] | p.[Asp134Gly]; [Ser299Pro] | p.[Pro155Leu]; [Ser21∗] | p.[Gln172∗]; [Gln172∗] | p.[Gln244∗]; [Gln244∗] | p.[?]; [?] | p.[Lys118∗]; [Lys118∗] | whole gene deletion |
| Demographics | ||||||||||
| Sex | F | M | M | M | M | F | M | F | F | M |
| Age at last follow-up | 28 years | 26 years | 3.5 months (died) | 3.5 months (died) | 2 years (died) | 8 years 9 months | 13 years 3 months | 7 years | 7 years | 5 years 6 months |
| Weight | 24 kg (−5 SD) | 26 kg (−5 SD) | NA | 3.3 kg (- 4 SD) | failure to thrive | 22 kg (8th centile) | 62 kg (85th centile) | 27.6 kg (50th centile) | normal | N/A |
| Height | 112 cm (−8 SD) | 128 cm (−7 SD) | NA | 50.3 cm (−5 SD) | short stature | 130.8 cm (46th centile) | 155 cm (25th centile) | 130 cm (90th centile) | normal | 105 cm (<3rd centile) |
| Consanguinity | – | – | – | – | – | – | + | + | – | + |
| Gestational/perinatal record | ||||||||||
| Fetal ultrasound | hydronephrosis | hydronephrosis | short limbs | N/A | severe IUGR | unremarkable | unremarkable | unremarkable | unremarkable | unremarkable |
| Gestation age | term | term | term | term | prematurity | term | term | term | term | term |
| Birth weight | N/A | N/A | 2180 g (−3.2 SD) | 2,000 g (−3.3 SD) | N/A | 2,760 g | 2,810 g (−1.6 SD) | 2,600 g (−2 SD) | 4,010 g (91st centile) | 3,000 g (50th centile) |
| Birth length | N/A | N/A | 40 cm (−5 SD) | 41.7 cm (−4.3 SD) | N/A | 50 cm at 6 weeks (−2.1 SD) | 49 cm (−1.3 SD) | normal | 49.5 cm (43rd centile) | 50 cm (−1.3 SD) |
| Skeletal radiograph | ||||||||||
| Spine anomaly | + | + | + | + | N/A | – | – | – | – | – |
| Metaphyseal anomaly | N/A | N/A | + | + | N/A | – | – | – | – | – |
| Epiphyseal anomaly | + | + | + | + | + | – | – | + (wrists) | – | – |
| Syndactyly | + | + | + | + | + | + | – | + | – | + |
| Brachydactyly/clinodactyly/camptodactyly | + | + | + | + | + | + | + | + | + | + |
| Others | ||||||||||
| Cardiac anomaly | – | – | + | – | + | + | – | – | – | + |
| Hydronephrosis | + | + | N/A | – | + | – | – | + | N/A | – |
| Intellectual disability/developmental delay | – | – | N/A | N/A | +b | +c | +d | +d | + | + |
M, male; F, female; N/A, not available; IUGR, intrauterine growth restriction.
Variants are named according to GenBank: NM_153332.4 or NC_000008.11.
Delayed motor milestones and speech, generalized hypotonia.
Global developmental delay, autism.
Intellectual disability, mild.
Although various digital abnormalities existed in all affected individuals (Figures 1E–1N and 2Q–2W), SEMD was only present in those with missense ERI1 variants (families 1–4) (Figures 2A–2P, Table 1). Additional clinical manifestations, including cardiac anomalies, developmental delay, and hydronephrosis, were found in some of the individuals (Table 1). Clinical summaries are provided in the supplemental note and pedigrees in Figure S1. A search of known monogenic skeletal disorders, including disease entities registered in the latest version of the international nosology and classification of genetic skeletal disorders,31 did not find any reports matching the characteristic SEMD that was observed in this study. We tentatively suggest the name SEMD, Guo-Campeau type for this phenotype.
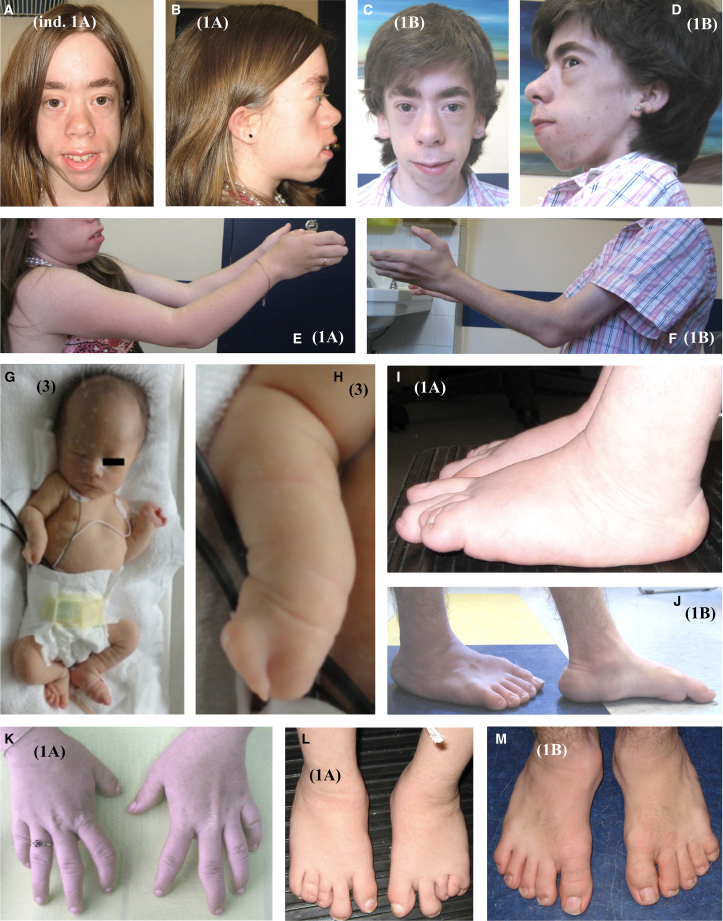
Figure 1.
Photographs of affected individuals
(A and B) Individual 1A. Note zygomatic hypoplasia, prominent alveolar processes of the maxilla and mandible, and small and low-set ears.
(C and D) Individual 1B. Note zygomatic hypoplasia and prominent alveolar processes of the maxilla and mandible.
(E and F) Maximal elbow extension for individuals 1A and 1B. Note distal camptodactyly in individual 1B.
(G and H) Individual 3, born at full term. Short stature with short limbs (G) and oligo-syndactyly of the right hand (H) and the right foot (G). The black box in (G) is used to increase anonymity.
(I and J) Lateral view of feet of individuals 1A (I) and 1B (J). Note short feet.
(K) Hands of individual 1A. Clinodactyly of the proximal interphalangeal joint of second right digit.
(L and M) Dorsal view of feet of individuals 1A (L) and 1B (M). Note short feet and 4-5 syndactyly.
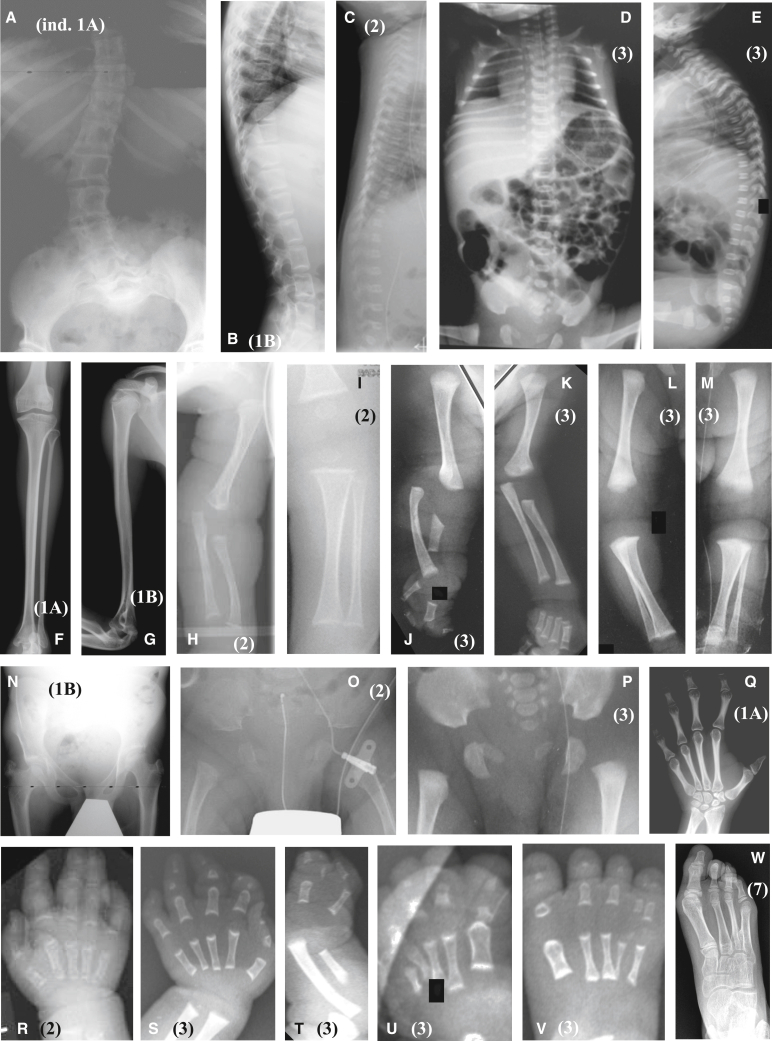
Figure 2.
Radiographs of affected individuals
(A) Lower spine of individual 1A with lumbar scoliosis.
(B) Lateral spine of individual 1B showing flattened vertebral bodies and irregular plates.
(C–E) Platyspondyly in individuals 2 (C) and 3 (D and E).
(F–M) Long tubular bones showing epi-metaphyseal dysplasia characterized by irregular epiphyses and frayed metaphyses in individuals 1A (F), 1B (G), 2 (H and I), and 3 (J–M). The right upper limb of individual 3 shows ulnar hypoplasia and radial bowing with elbow dislocation (J).
(N–P) Hypoplasia of pelvis and epi-metaphyseal dysplasia of femurs in individuals 1B (N), 2 (O), and 3 (P). Note right coxa vara and left coxa valga (N), iliac hypoplasia and delayed pubic ossification (O and P).
(Q) Right hand of individual 1A. Note single thumb phalanx, ulnar deviation of the second finger at proximal interphalangeal joint, and narrow metacarpal bones.
(R) Left hand of individual 2 showing brachy-syndactyly of the third and fourth fingers.
(S–V) Hands and feet of individual 3. The left hand (S) shows brachydactyly with hypoplasia of all middle phalanges, the first proximal and distal phalanges, and the second and third distal phalanges as well as ulnar clinodactyly of the second and third fingers and radial clinodactyly of the fourth and fifth fingers. The proximal end of the proximal phalanges and distal ends of the metacarpals are conspicuously cupped. The right hand shows oligo-syndactyly (split hand) (T). Both feet show postaxial syndactyly and the third and fourth metatarsal bases may be fused (U and V). The right fourth metatarsal is hypoplastic (V).
(W) Right foot of individual 7. Note absence of the fifth ray, proximal fusion of the third and fourth metatarsal bones, and widened fourth metatarsus. The black boxes in (J), (L), and (U) mask arrows previously used in clinical discussions.
In contrast, three individuals with bi-allelic ERI1 null mutations (families 5–7) showed a much milder skeletal phenotype, which affects the patterning of digital bones but without any chondrodysplastic feature such as SEMD (Table 1). The prominent clinical manifestation of these individuals was intellectual disability (Table 1), consistent with the previous individual with homozygous 284 kb deletion reported by the Mégarbané’s group.10
The ERI1 variants exhibit loss of 3′-to-5′ exoribonuclease activity
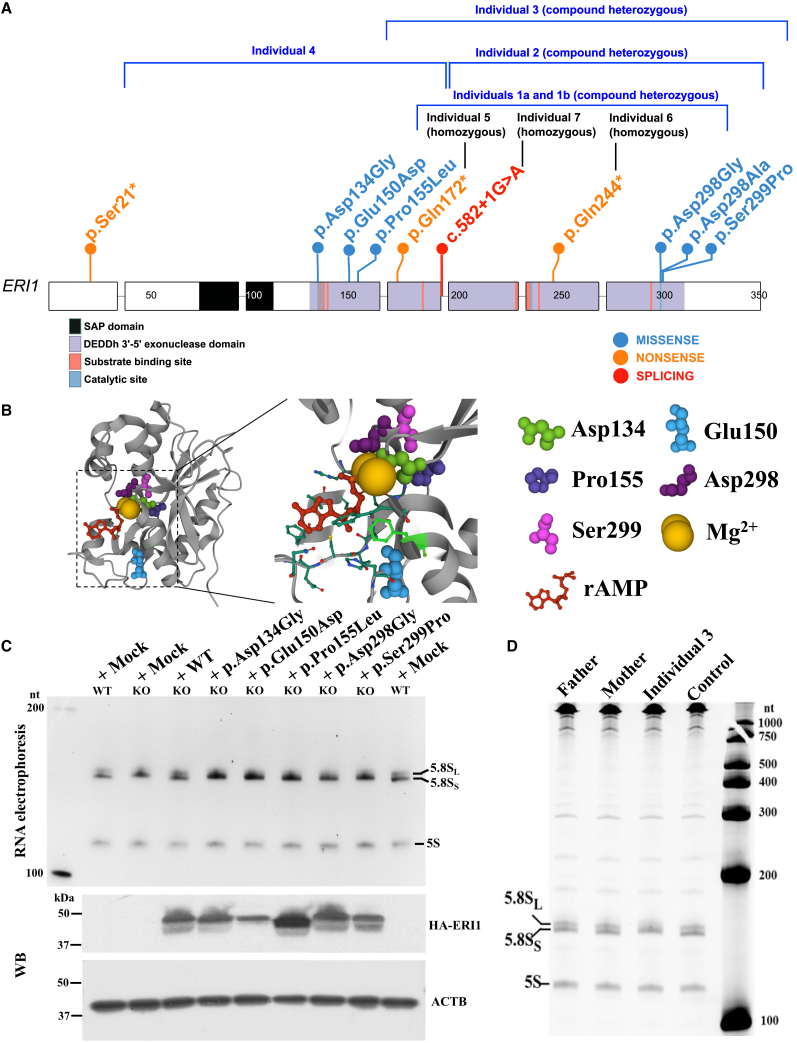
ERI1 includes two functional domains: the amino-terminal SAP (SAF-A/B, Acinus, and PIAS) domain and the carboxy-terminal 3′ exonuclease domain (Figure 3A). The structure of the exonuclease domain complexed with riboadenine-5′-monophosphate (rAMP) is determined in the presence of Mg2+ ions (Figure 3B).32 The six missense variants, p.Asp134Gly, p.Glu150Asp, p.Pro155Leu, p.Asp298Gly, p.Asp298Ala, and p.Ser299Pro, identified in our study are all located in the carboxy-terminal 3′ exonuclease domain of ERI1 (Figure 3A). A 3D structure of ERI1 indicates that the residues corresponding to these missense variants are parts of the active site of the nuclease domain and are required to coordinate the two Mg2+ ions (Figure 3B), thus suggesting their key roles for ERI1 enzyme activity. c.582+1G>A was a canonical splice-disrupting variant. The location of the nonsense variants p.Ser21∗, p.Gln172∗, and p.Gln244∗ suggests that all of them are subjected to nonsense-mediated mRNA decay.
Figure 3.
Functional analysis of ERI1 pathogenic variants
(A) Schematic overview of ERI1 showing the position of the variants identified in the individuals. ERI1 includes two functional domains: amino-terminal SAP (SAF-A/B, Acinus, and PIAS) domain and carboxy-terminal 3′ exonuclease domain.
(B) 3D structure of ERI1 with annotated residues Asp134, Glu150, Pro155, Asp298, and Ser299 generated via UniProKB (https://www.uniprot.org/uniprot/Q8IV48). PDB: 1ZBU. rAMP, riboadenine-5′-monophosphate.
(C) The upper panel shows ethidium bromide-stained RNA from the wild type (WT) and ERI1 knockout (KO) HeLa cells. Nt, nucleotide. WT, p.Asp134Gly, p.Glu150Asp, p.Pro155Leu, p.Asp298Gly, and p.Ser299Pro ERI1 were produced in the KO cells to rescue the 3′-to-5′ exoribonuclease activity of ERI1 in catalyzing 5.8S ribosomal RNA (rRNA) processing. The lower panel shows western blot of whole-cell lysates.
(D) Ethidium bromide-stained RNA from the lymphoblastoid cell lines of the affected individual 3, his parents, and a healthy control. 5.8SL, long form of 5.8S rRNA; 5.8SS, short form of 5.8S rRNA.
Eri1 is a 3′-to-5′ exoribonuclease that catalyzes 5.8S rRNA processing.7,9 To study a potential deleterious effect of these missense ERI1 variants, we generated an ERI1 KO HeLa cell line (Figure S2) and demonstrated that ERI1 was required for proper 5.8 rRNA 3′ end formation also in human cells (Figures 3C and S2F), consistent with the previous findings in mice, worms, and fission yeast.7,9 Then we attempted to rescue defective 5.8S rRNA processing by generating ERI1 from the WT construct and the missense variant constructs in KO HeLa cells. While WT ERI1 efficiently catalyzed conversion of the abnormal 5.8S rRNA to its normal size, all of the variant ERI1s failed to do so (Figure 3C). Consistently, electrophoresis of total RNA extracted from the family 3-derived lymphoblastoid cells showed that the affected individual, but not his parents or an unrelated unaffected individual, exhibited the same 5.8S rRNA processing defect as the KO HeLa cells (Figure 3D). Thus, both in vitro and in vivo evidence indicates that the ERI1 missense variants cause a loss of function of the 3′-to-5′ exoribonuclease activity of ERI1.
Effect of ERI1 deficiency on abundance of replication-dependent histone mRNAs and miRNAs
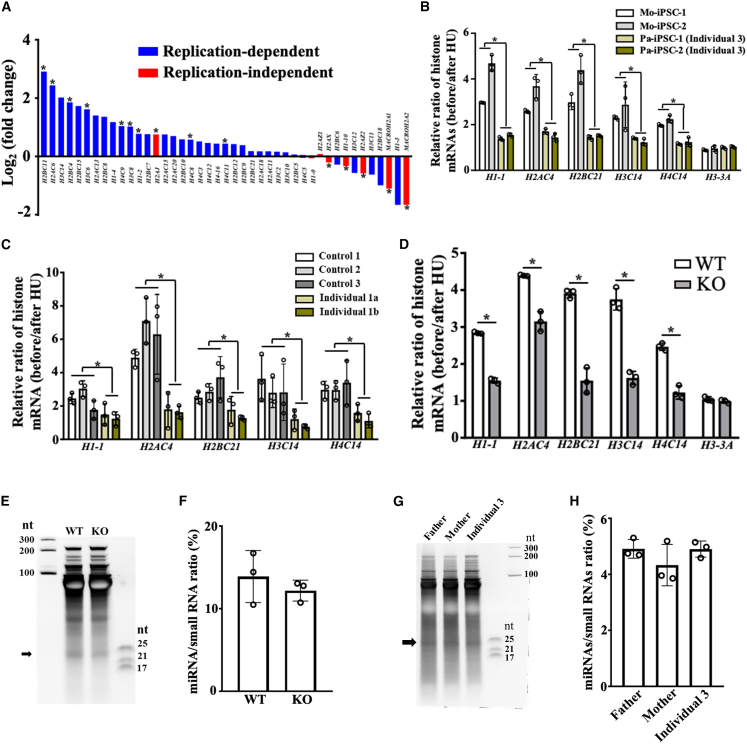
Besides 5.8S rRNA processing, Eri1 regulates the abundance of histone mRNAs and miRNAs in mouse cells.33 In mouse embryonic fibroblasts, loss of Eri1 leads to excessive accumulation of replication-dependent histone mRNAs.8 To assess the corresponding function in human cells, we analyzed histone mRNA levels in the mother- and affected-individual-derived iPSCs from family 3 by using RNA-seq data. Indeed, the replication-dependent histone mRNAs showed an overall increased abundance in the affected-individual-derived iPSCs compared to the replication-independent histone mRNAs (Figure 4A), in accordance with the previous findings in mouse cells.8 This effect was also evident in the ERI1 KO HeLa cells and affected-individual-derived iPSCs and fibroblasts treated with hydroxyurea (Figures 4B–4D), which induces degradation of replication-dependent histone mRNAs similar to their physiologic regulation at the end of S phase.8 These findings confirm the regulatory role of ERI1 in this class of mRNAs as shown in mouse cells8 and indicate a decreased degradation of replication-dependent histone mRNAs in affected individuals.
Figure 4.
Effect of ERI1 deficiency on histone mRNAs and miRNAs
(A) Histone mRNA levels of the family 3-derived iPSCs in the process of chondrogenic differentiation (day 8). Both replication-dependent and replication-independent histone genes are ranked according to the fold change (FC) of the differentially expressed genes in Mo-iPSC-1 versus Pa-iPSC-1. ∗p < 0.05. n = 3 biologically independent samples. Statistical significance was assessed via two-sided t test. p value was adjusted by false discovery rate (FDR).
(B–D) qPCR for histone mRNAs before and after hydroxyurea (HU) treatment for 45 min. All markers are replication-dependent histone genes except H3-3A.
(B) Family 3-derived iPSCs.
(C) Fibroblasts from three healthy control individuals and the two affected individuals in family 1.
(D) Wild type (WT) and knock-out (KO) HeLa cells. The data in (B)–(D) indicate mean ± SD. n = 3 biological replicates. Statistical test was assessed via multiple t tests. ∗p < 0.05.
(E–H) SYBR Gold-stained small RNAs from the wild type (WT) and ERI1 knockout (KO) HeLa cells (E) and family 3-derived lymphoblastoid cells (G). nt, nucleotide. The arrows show single-strand RNAs between 21 and 25 nt, corresponding to the abundance of miRNAs. Quantification of the miRNA to total small RNA (<300 nt) ratio in HeLa cells (F) and lymphoblastoid cells (H). The data indicate mean ± SD. p = 0.44 and 0.33. n = 3 biological replicates. Statistical significance was assessed via two-sided t test and one-way ANOVA.
Additionally, conditional deletion of Eri1 in mouse natural killer (NK) and T cells displayed a global, sequence-independent increase in miRNA abundance.6 We electrophoresed total small RNAs shorter than 300 nucleotides (nts) extracted from WT and ERI1 KO HeLa cells and affected-individual-derived lymphoblastoid cell lines to evaluate the proportion of miRNAs (Figures 4E and 4G). We enhanced the visualization of small RNAs by using SYBR Gold Nucleic Acid Gel Stain.34 As a result, a smear of bands corresponding to single-strand RNAs (ssRNAs) between 21 and 25 nts was detected; however, the overall abundance of these small RNA species did not change in ERI1 KO HeLa cells or affected-individual-derived lymphoblastoid cells (Figures 4F and 4H).
Impaired chondrogenesis in affected-individual-derived iPSCs
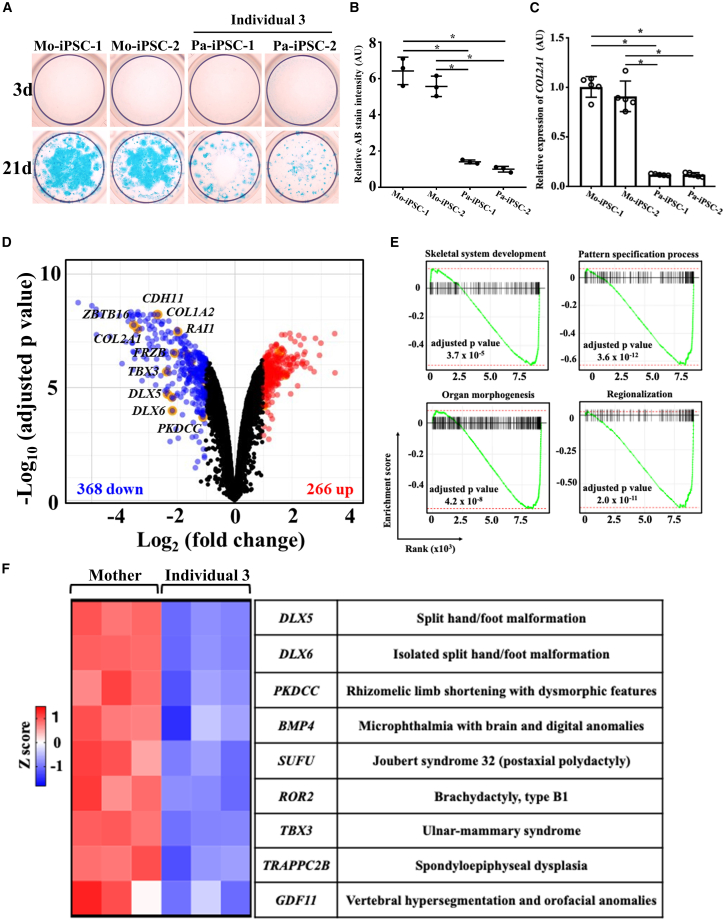
To examine possible roles of ERI1 in chondrogenesis that could then explain the etiology of the individuals’ skeletal phenotypes, we established and verified four iPSC lines by using affected-individual-derived lymphoblastoid cell lines (Figure S3), which were generated from peripheral blood lymphocytes of an affected individual and his mother from family 3. The iPSC lines were differentiated toward chondrocytes as described previously.29 While Alcian blue (AB) staining of sulfated glycosaminoglycan accumulated in the mother-derived lines at day 21, significantly decreased AB staining was found in the affect-individual-derived lines (Figures 5A and 5B). The expression of the chondrocyte marker COL2A1 was also decreased in the affected-individual-derived lines, as compared to its expression in the mother-derived lines (Figure 5C). We then subjected the differentiating iPSCs at day 8 (which correspond to mesodermal cells29) to RNA-seq. We identified 634 differentially expressed genes (Figure 5D). A gene set enrichment analysis identified several Gene Ontology (GO) terms, including “skeletal system development” and “pattern specification process” (Figure 5E), in which the genes were mostly downregulated in the affected-individual-derived cells. The downregulated genes included DLX5, DLX6, PKDCC, BMP4, SUFU, ROR2, TBX3, TRAPPC2B, and GDF11, which are associated with skeletal dysplasias,35,36,37,38,39,40,41 thereby uncovering disease-relevant changes in mesodermal differentiation of cells from affected individuals (Figure 5F). These skeletal dysplasias have defective development in the short tubular bones, such as split hand/foot malformation (MIM: 183600) and brachydactyly, type B1 (MIM: 113000), and in the long tubular bones and spine, such as spondyloepiphyseal dysplasia (Figure 5F), which are similar to the skeletal phenotypes of affected individuals described here. These results indicate that affected-individual-derived iPSCs with bi-allelic missense variants are impaired in their ability to undergo chondrogenesis, suggesting that defects in both digital patterning and chondrogenesis are caused by missense variants.
Figure 5.
Chondrogenic differentiation of the affected-individual-derived induced pluripotent stem cells (iPSCs)
(A) Alcian blue (AB) staining for the chondrocytic nodules induced from the healthy mother iPSCs (Mo-iPSC-1 and -2) and affected-individual-derived iPSCs (Pa-iPSC-1 and -2) in family 3.
(B) Quantification of AB staining; n = 3 biologically independent samples. ∗p < 0.05. Error bars represent SD.
(C) qPCR for COL2A1; n = 5 biologically independent samples. ∗p < 0.05. AU, arbitrary unit. The data in (B) and (C) indicate mean ± SD. Statistical test was assessed via one-way ANOVA.
(D) Volcano plot of RNA-seq data for the differentiating iPSCs at day 8 showing differentially expressed genes (DEGs) in Mo-iPSC-1 versus Pa-iPSC-1 (n = 3 biologically independent samples) with respect to fold change (FC) and significance (adjusted p value). Each dot represents an individual gene. Blue dots represent significant downregulated DEGs (FC < 0.5) and red dots represent significant upregulated DEGs (FC > 2). Statistical significance was assessed via moderated t test in limma. p value is adjusted by false discovery rate.
(E) Gene set enrichment analysis for RNA-seq data in (D). The genes belonging to the corresponding GO terms are enriched at the bottom of the entire ranked list of genes.
(F) Heatmap of DEGs associated causally with human skeletal dysplasia.
Eri1 KO mice partially recapitulate skeletal dysplasia caused by human ERI1 pathogenic variants
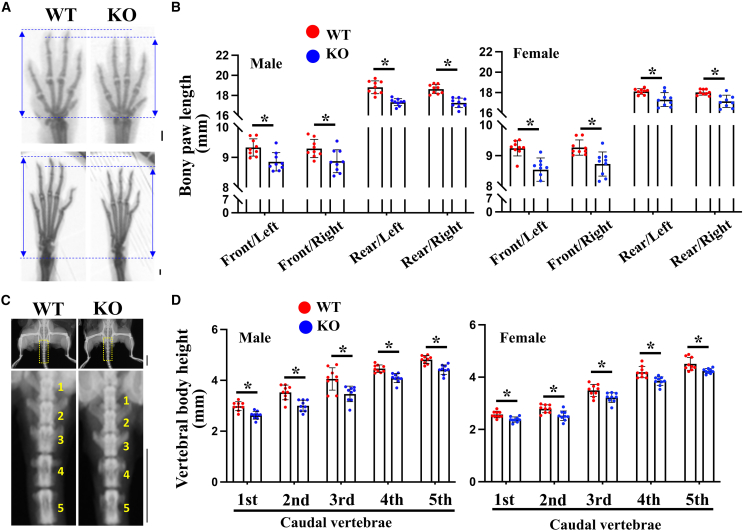
Eri1 KO mice on the C57BL/6 inbred strain background show a growth defect with reduced body size and neonatal mortality.7 Neonatal lethality was rescued by breeding Eri1 KO mice on outbred strain backgrounds.7 To test potential phenotypic similarities between the affected individuals and the KO mice, we evaluated the morphological changes of the Eri1 KO mice skeleton on an NMRI × C57BL/6 mixed background. The Eri1 KO mice showed brachydactyly (Figures 6A and 6B), which was also characteristic for the affected individuals with ERI1 variants (Figures 2Q–2W). The examination of X-rays and histological stain did not show obvious change in the epimetaphyseal regions of the long tubular bones of the Eri1 KO mice (Figure S4). Although the Eri1 KO mice showed platyspondyly (Figures 6C and 6D), which was also characteristic for the affected individuals with ERI1 missense variants (Figures 2A–2E), such a phenotype was mild in the KO mice, perhaps being secondary to the reduced body size. Therefore, the skeletal phenotypes of Eri1 KO mice were more similar to the affected individuals with null variants than those with missense variants.
Figure 6.
Skeletal dysplasia of Eri1 knockout (KO) mice
(A) X-ray images of paws of the wild type (WT) and Eri1 KO mice at 16 weeks. Scale bar, 1 mm.
(B) Quantitative analysis of the paw length shown in (A) (n = 9 mice). ∗p < 0.05. The above quantitative data indicate mean ± SD and statistical significance was assessed via two-sided t test. Error bars represent SD.
(C) X-ray images of the WT and Eri1 KO mice at 16 weeks. The areas highlighted by the dotted box in the upper panels are magnified in the lower panels. Scale bar, 1 cm.
(D) Measurement of the height of the caudal vertebral bodies shown in (C) (n = 9 mice). ∗p < 0.05.
Discussion
In this study, we describe a type of SEMD by characterizing the phenotypes caused by different types of bi-allelic ERI1 variants. The SEMD was present in the five individuals with at least one missense variant (Table 1). In contrast, three individuals with ERI1 null mutations and the Eri1 KO mice showed a much milder skeletal phenotype without any evidence for SEMD, consistent with the two individuals reported previously, who had homozygous a 284 kb deletion and p.Lys118∗. Notably, of the five individuals with SEMD, three died within 2 years after birth, suggesting missense variants lead to a poor prognosis.
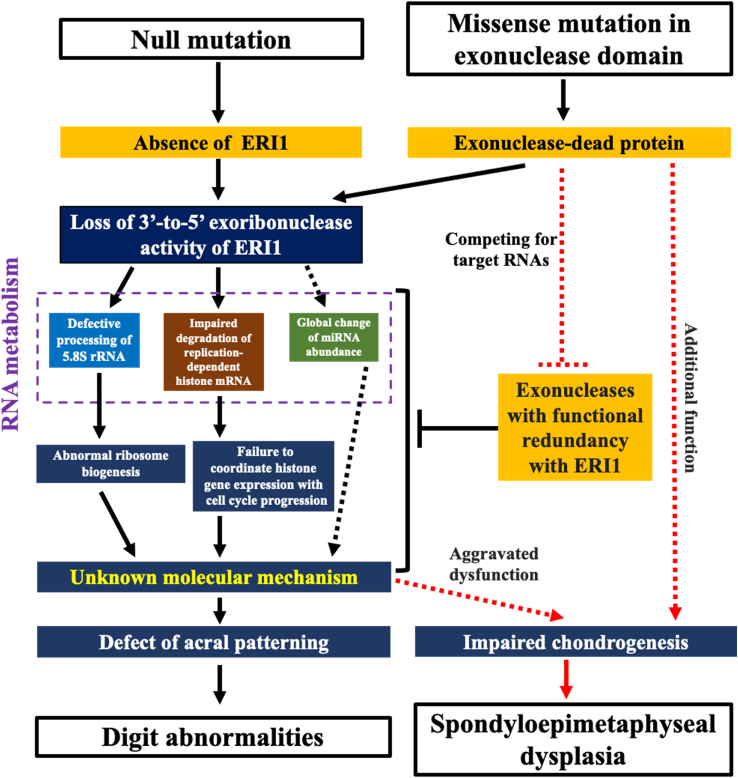
The phenomenon that more severe clinical phenotypes are associated with missense variants rather than null variants is frequently observed in some dominant disorders, such as osteogenesis imperfecta caused by COL1A1 and COL1A2 mutations,42 but not reported in recessive disorders. All missense variants in affected individuals are located in the carboxy-terminal 3′ exonuclease domain without disrupting the sequence of the amino-terminal SAP domain, which is an RNA-binding motif. These exonuclease-dead proteins may compete for the target RNA molecules with other exonucleases that have functional redundancy with ERI1, staying bound to those RNA molecules and thus leading to a more severe phenotype in skeleton (Figure 7). Alternatively, these exonuclease-dead proteins may impair the chondrogenesis by an additional mechanism (Figure 7). The genotype-phenotype association needs to be verified further by finding more affected individuals and comparing the phenotypic difference between the KO mice and the knockin mice to be generated with the individual-specific missense variants.
Figure 7.
Hypothesis of the pathological mechanism underlying the phenotypic dichotomy between null and missense variants of ERI1
Both null and missense mutations of ERI1 cause loss of 3′-to-5′ exoribonuclease activity of ERI1, leading to defective processing of 5.8S rRNA and impaired degradation of replication-dependent histone mRNA in humans, which interrupt diverse cellular processes including ribosome biogenesis and coordination of histone gene expression with cell cycle progression. These defects singly or jointly alter gene expression profiling that regulates acral patterning, thus inducing digit abnormalities. ERI1 missense mutations also produce exonuclease-dead proteins, which may compete with other exonucleases having functional redundancy with ERI1, thus aggravating the dysfunction of ERI1-mediated RNA metabolism. The aggravated dysfunction or additional unknown effects lead to impaired chondrogenesis that causes spondyloepimetaphyseal dysplasia, finally contributing to the phenotypic dichotomy between null and missense variants of ERI1. Solid arrow, causal connection; dashed arrow, hypothetical connection; red line, additional effects of exonuclease-dead ERI1 to the loss of the exoribonuclease activity.
Eri1 is a 3′-to-5′ exoribonuclease, whose functions generally show evolutionary conservation in the processing of 5.8S rRNA and histone mRNAs and species-specific differences in the regulation of small RNA pathways that include endogenous siRNAs and miRNAs.9,33,43 The 5.8S rRNA 3′ end forms a duplex reminiscent of pre-miRNAs by base pairing with the 5′ end of the 28S rRNA, which leaves a 3′ ssRNA overhang processed by Eri1 into a duplex with 1–2 nt 3′ overhang.7,9 Our study demonstrated that ERI1 processes 5.8S rRNA in humans as it does in other species. Although the biological significance of the ERI1-mediated 5.8S rRNA 3′ trimming remains unknown, the biological process is conserved from yeast to humans, suggesting its importance in ribosome biogenesis, a complex endeavor that involves coordination of three RNA polymerases, approximately 80 ribosomal proteins (RPs), 70 small nucleolar RNAs, and several rRNAs.44
Variants in ribosomal proteins and regulators of ribosome biogenesis lead to ribosomopathies, a group of disorders that show developmental defects in craniofacial bones and long and short tubular bones, causing skeletal dysplasias such as Treacher Collins syndrome 1 (MIM: 154500), postaxial acrofacial dysostosis, Shwachman-Diamond syndrome 1 (MIM: 260400), Roberts syndrome, and cartilage hair hypoplasia (MIM: 250250).45,46,47,48,49 The skeletal phenotypes of these disorders include spondylo-metaphyseal dysplasia and digit deformities, showing similarities with affected individuals reported here. Ribosomopathies are also characterized by hematologic phenotypes such as macrocytic anemia in Diamond-Blackfan anemia 1 (MIM: 105650) and congenital neutropenia in Shwachman-Diamond-like syndrome.50,51,52 Macrocytic anemia and congenital neutropenia were also evident in individual 3 and the Eri1 KO mice (Figures S5 and S6). Thus, both the biological function of ERI1 and the clinical manifestations suggest existence of a ribosomopathy in the individuals reported here. The known genes associated with ribosomopathies cover functions in methylation and cleavage of 47S precursor rRNA, 18S rRNA maturation, and assembly of ribosomal subunits.53 The defective processing of 5.8S rRNA in our cohort would provide a link that associates human disease with ribosome biogenesis, thus enriching the present list of ribosomopathies.
Eri1 participates in fast degradation of replication-dependent histone mRNAs in mice by binding to the oligouridylated histone mRNA 3′ end and trimming two nucleotides at the end of S phase, similar to its role in 5.8S rRNA maturation.8 Marked accumulation of replication-dependent histone mRNAs were observed in our affected-individual-derived cells and ERI1 KO HeLa cells as found in the Eri1 KO mouse cells, indicating the function of histone mRNA exonuclease of ERI1 is also conserved in humans. High levels of histone mRNAs are closely linked to DNA replication so that numerous histones are produced in S phase to accommodate DNA packaging into nucleosomes. Histone mRNA abundance then falls rapidly in G2 by fast degradation. Although the biological effects of excessive replication-dependent histone mRNAs remain unknown in mammals, failure to coordinate histone gene expression with cell-cycle progression results in genomic instability and cell-cycle arrest in yeast.54 Our study suggests that the defective histone mRNA decay may disrupt skeletal development singly or in combination with the defective 5.8S rRNA maturation in the affected individuals.
Previous studies using Eri1 KO mice reported that Eri1 loss causes a global, sequence-independent increase in mature miRNA abundance in NK and T cells.6 Unexpectedly, we did not detect a global increase in the abundance of 21–25 nt ssRNAs in ERI1 KO HeLa cells and affected-individual-derived lymphoblastoid cells. Eri1 is highly expressed in mouse spleen and thymus compared to other tissues.7 NK and T lymphocytes upregulate Eri1 expression to high levels upon activation, thus suggesting the Eri1 effects on miRNA turnover may be cell/context-dependent.6 Alternatively, the regulatory role of ERI1 in miRNA homeostasis may not be completely conserved between mice and humans.
The multiple RNA metabolic pathways regulated by Eri1 make the deeply conserved exoribonuclease play an extremely complex role in the crosstalk and co-regulation of diverse cellular processes. Our study establishes a causal link between ERI1 deficiency and human skeletal dysplasia and reveals the evolutionarily conserved functions of ERI1 in RNA metabolic pathways, which associate 5.8S rRNA maturation and histone mRNA decay with the regulation of bone morphogenesis (Figure 7), thus leading to further exploration of the roles of ERI1-mediated RNA metabolism in skeletal development. Our study reveals a phenotypic dichotomy between null and missense variants of ERI1, thus serving as a precedent for a nuanced form of mechanistic heterogeneity for recessive conditions in humans (Figure 7).
Acknowledgments
Author contributions
The study was designed by L.G., S.I., and P.M.C. The individuals were recruited by N.N., H.K., J.C.S., N.D.N., S.B., A.J., I.H., T.B., C.B., F.A., R.M., G.C., B.W., A.M., T.Y., G.N., J.A.R., S.I., and P.M.C. The disease was evaluated by L.G., T.Y., G.N., S.I., and P.M.C. The experiments were performed or supervised by L.G., S.S., J.R., H.K., J.Y.X., N.R., Z.W., Z.J.W., H.O., M.F.T., N. Matsumoto, N. Miyake, M.K.A., S.M., G.Y., B.W., M.H.d.A., and V.H. The data were analyzed by L.G., S.S., J.Y.X., J.W., and Y.M.Z. The draft was written by L.G., S.I., and P.M.C. All authors contributed to revising the manuscript.
Declaration of interests
The Department of Molecular and Human Genetics at Baylor College of Medicine receives revenue from clinical genetic testing completed at Baylor Genetics Laboratories.
Published: June 22, 2023
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.ajhg.2023.06.001.
Contributor Information
Long Guo, Email: longguo601@gmail.com.
Philippe M. Campeau, Email: p.campeau@umontreal.ca.
Web resources
bioinfokit, https://pypi.org/project/bioinfokit/
E-CRISP, http://www.e-crisp.org/E-CRISP/
ExpressAnalyst, https://www.expressanalyst.ca/
featureCounts, https://subread.sourceforge.net/
GeneMatcher https://genematcher.org/
gnomAD population database, https://gnomad.broadinstitute.org/
Human Gene Mutation Database, https://portal.biobase-international.com/hgmd/pro/start.php
Human Genome Variation Society guidelines, http://www.hgvs.org
limma https://bioconductor.org/packages/release/bioc/html/limma.html
OMIM, https://omim.org/
UniProKB, https://www.uniprot.org/uniprot/
Supplemental information
Data and code availability
The RNA-seq data have been deposited in NCBI GEO with the accession code GEO: GSE206412. The individuals’ genomic data obtained by exome or genome sequencing are not publicly available because study participants did not give full consent for releasing data publicly. These data can be accessed under the condition that a joint research plan is made by the researchers and approved by the ethics committees of the participating institutions. All the other data supporting the findings of this study are included within the article or the supplemental information.
References
- 1.Zuo Y., Deutscher M.P. Exoribonuclease superfamilies: Structural analysis and phylogenetic distribution. Nucleic Acids Res. 2001;29:1017–1026. doi: 10.1093/nar/29.5.1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Simmer F., Tijsterman M., Parrish S., Koushika S.P., Nonet M.L., Fire A., Ahringer J., Plasterk R.H.A. Loss of the putative RNA-directed RNA polymerase RRF-3 makes C. Elegans hypersensitive to RNAi. Curr. Biol. 2002;12:1317–1319. doi: 10.1016/s0960-9822(02)01041-2. [DOI] [PubMed] [Google Scholar]
- 3.Gu W., Shirayama M., Conte D., Vasale J., Batista P.J., Claycomb J.M., Moresco J.J., Youngman E.M., Keys J., Stoltz M.J., et al. Distinct Argonaute-Mediated 22G-RNA Pathways Direct Genome Surveillance in the C. elegans Germline. Mol. Cell. 2009;36:231–244. doi: 10.1016/j.molcel.2009.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Han T., Manoharan A.P., Harkins T.T., Bouffard P., Fitzpatrick C., Chu D.S., Thierry-Mieg D., Thierry-Mieg J., Kim J.K. 26G endo-siRNAs regulate spermatogenic and zygotic gene expression in Caenorhabditis elegans. Proc.Natl.Acad.Sci.USA. 2009;106:18674–18679. doi: 10.1073/pnas.0906378106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bühler M., Verdel A., Moazed D. Tethering RITS to a Nascent Transcript Initiates RNAi- and Heterochromatin-Dependent Gene Silencing. Cell. 2006;125:873–886. doi: 10.1016/j.cell.2006.04.025. [DOI] [PubMed] [Google Scholar]
- 6.Thomas M.F., Abdul-Wajid S., Panduro M., Babiarz J.E., Rajaram M., Woodruff P., Lanier L.L., Heissmeyer V., Ansel K.M. Eri1 regulates microRNA homeostasis and mouse lymphocyte development and antiviral function. Blood. 2012;120:130–142. doi: 10.1182/blood-2011-11-394072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ansel K.M., Pastor W.A., Rath N., Lapan A.D., Glasmacher E., Wolf C., Smith L.C., Papadopoulou N., Lamperti E.D., Tahiliani M., et al. Mouse Eri1 interacts with the ribosome and catalyzes 5.8S rRNA processing. Nat. Struct. Mol. Biol. 2008;15:523–530. doi: 10.1038/nsmb.1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hoefig K.P., Rath N., Heinz G.A., Wolf C., Dameris J., Schepers A., Kremmer E., Ansel K.M., Heissmeyer V. Eri1 degrades the stem-loop of oligouridylated histone mRNAs to induce replication-dependent decay. Nat. Struct. Mol. Biol. 2013;20:73–81. doi: 10.1038/nsmb.2450. [DOI] [PubMed] [Google Scholar]
- 9.Gabel H.W., Ruvkun G. The exonuclease ERI-1 has a conserved dual role in 5.8S rRNA processing and RNAi. Nat. Struct. Mol. Biol. 2008;15:531–533. doi: 10.1038/nsmb.1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Choucair N., Rajab M., Mégarbané A., Chouery E. Homozygous microdeletion of the ERI1 and MFHAS1 genes in a patient with intellectual disability, limb abnormalities, and cardiac malformation. Am. J. Med. Genet. 2017;173:1955–1960. doi: 10.1002/ajmg.a.38271. [DOI] [PubMed] [Google Scholar]
- 11.Hoxha V., Aliu E. ERI1: A case report of an autosomal recessive syndrome associated with developmental delay and distal limb abnormalities. Am. J. Med. Genet. 2023;191:64–69. doi: 10.1002/ajmg.a.62987. [DOI] [PubMed] [Google Scholar]
- 12.Ravera S., Dufour C., Cesaro S., Bottega R., Faleschini M., Cuccarolo P., Corsolini F., Usai C., Columbaro M., Cipolli M., et al. Evaluation of energy metabolism and calcium homeostasis in cells affected by Shwachman-Diamond syndrome. Sci. Rep. 2016;6:25441. doi: 10.1038/srep25441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Inflammatory Signature, Oxidative Stress, and DNA Damage Response in DBA Pathogenesis. Blood. 2017;130:2452. [Google Scholar]
- 14.Zambetti N.A., Ping Z., Chen S., Kenswil K.J.G., Mylona M.A., Sanders M.A., Hoogenboezem R.M., Bindels E.M.J., Adisty M.N., Van Strien P.M.H., et al. Mesenchymal Inflammation Drives Genotoxic Stress in Hematopoietic Stem Cells and Predicts Disease Evolution in Human Pre-leukemia. Cell Stem Cell. 2016;19:613–627. doi: 10.1016/j.stem.2016.08.021. [DOI] [PubMed] [Google Scholar]
- 15.Hermanns P., Bertuch A.A., Bertin T.K., Dawson B., Schmitt M.E., Shaw C., Zabel B., Lee B. Consequences of mutations in the non-coding RMRP RNA in cartilage-hair hypoplasia. Hum. Mol. Genet. 2005;14:3723–3740. doi: 10.1093/hmg/ddi403. [DOI] [PubMed] [Google Scholar]
- 16.Brunet T., Jech R., Brugger M., Kovacs R., Alhaddad B., Leszinski G., Riedhammer K.M., Westphal D.S., Mahle I., Mayerhanser K., et al. De novo variants in neurodevelopmental disorders-experiences from a tertiary care center. Clin. Genet. 2021;100:14–28. doi: 10.1111/cge.13946. [DOI] [PubMed] [Google Scholar]
- 17.Yang Y., Muzny D.M., Xia F., Niu Z., Person R., Ding Y., Ward P., Braxton A., Wang M., Buhay C., et al. Molecular findings among patients referred for clinical whole-exome sequencing. JAMA. 2014;312:1870–1879. doi: 10.1001/jama.2014.14601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xue J.Y., Grigelioniene G., Wang Z., Nishimura G., Iida A., Matsumoto N., Tham E., Miyake N., Ikegawa S., Guo L. <scp> SLC4A2 </scp> Deficiency Causes a New Type of Osteopetrosis. J. Bone Miner. Res. 2022;37:226–235. doi: 10.1002/jbmr.4462. [DOI] [PubMed] [Google Scholar]
- 19.Guo L., Iida A., Bhavani G.S., Gowrishankar K., Wang Z., Xue J.Y., Wang J., Miyake N., Matsumoto N., Hasegawa T., et al. Deficiency of TMEM53 causes a previously unknown sclerosing bone disorder by dysregulation of BMP-SMAD signaling. Nat. Commun. 2021;12:2046–2113. doi: 10.1038/s41467-021-22340-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guo L., Elcioglu N.H., Karalar O.K., Topkar M.O., Wang Z., Sakamoto Y., Matsumoto N., Miyake N., Nishimura G., Ikegawa S. Dysosteosclerosis is also caused by TNFRSF11A mutation. J. Hum. Genet. 2018;63:769–774. doi: 10.1038/s10038-018-0447-6. [DOI] [PubMed] [Google Scholar]
- 21.Guo L., Bertola D.R., Takanohashi A., Saito A., Segawa Y., Yokota T., Ishibashi S., Nishida Y., Yamamoto G.L., Franco J.F.d.S., et al. Bi-allelic CSF1R Mutations Cause Skeletal Dysplasia of Dysosteosclerosis-Pyle Disease Spectrum and Degenerative Encephalopathy with Brain Malformation. Am. J. Hum. Genet. 2019;104:925–935. doi: 10.1016/j.ajhg.2019.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stavropoulos D.J., Merico D., Jobling R., Bowdin S., Monfared N., Thiruvahindrapuram B., Nalpathamkalam T., Pellecchia G., Yuen R.K.C., Szego M.J., et al. Whole Genome Sequencing Expands Diagnostic Utility and Improves Clinical Management in Pediatric Medicine. NPJ Genom. Med. 2016;1:15012. doi: 10.1038/npjgenmed.2015.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cong L., Ran F.A., Cox D., Lin S., Barretto R., Habib N., Hsu P.D., Wu X., Jiang W., Marraffini L.A., Zhang F. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Humbert J., Salian S., Makrythanasis P., Lemire G., Rousseau J., Ehresmann S., Garcia T., Alasiri R., Bottani A., Hanquinet S., Beaver E., Heeley J., Smith A.C.M., Berger S.I., Antonarakis S.E., Yang X.J., Côté J., Campeau P.M. De Novo KAT5 Variants Cause a Syndrome with Recognizable Facial Dysmorphisms, Cerebellar Atrophy, Sleep Disturbance, and Epilepsy. Am. J. Hum. Genet. 2020;107:564–574. doi: 10.1016/j.ajhg.2020.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fuchs H., Gailus-Durner V., Adler T., Pimentel J.A.A., Becker L., Bolle I., Brielmeier M., Calzada- Wack J., Dalke C., Ehrhardt N., et al. The German Mouse Clinic: A Platform for Systemic Phenotype Analysis of Mouse Models. Curr. Pharm. Biotechnol. 2009;10:236–243. doi: 10.2174/138920109787315051. [DOI] [PubMed] [Google Scholar]
- 26.Fukushima Y., Ohashi H., Wakui K., Nishida T., Oh-Ishi T. A rapid method for starting a culture for the establishment of Epstein-Barr virus-transformed human lymphoblastoid cell lines. J. Hum. Genet. 1992;37:149–150. doi: 10.1007/BF01899737. [DOI] [PubMed] [Google Scholar]
- 27.Kumar S., Curran J.E., Espinosa E.C., Glahn D.C., Blangero J. Highly efficient induced pluripotent stem cell reprogramming of cryopreserved lymphoblastoid cell lines. J. Biol. Methods. 2020;7:e124. doi: 10.14440/jbm.2020.296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Okita K., Matsumura Y., Sato Y., Okada A., Morizane A., Okamoto S., Hong H., Nakagawa M., Tanabe K., Tezuka K.I., et al. A more efficient method to generate integration-free human iPS cells. Nat. Methods. 2011;8:409–412. doi: 10.1038/nmeth.1591. [DOI] [PubMed] [Google Scholar]
- 29.Yamashita A., Morioka M., Yahara Y., Okada M., Kobayashi T., Kuriyama S., Matsuda S., Tsumaki N. Generation of scaffoldless hyaline cartilaginous tissue from human iPSCs. Stem Cell Rep. 2015;4:404–418. doi: 10.1016/j.stemcr.2015.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shukunami C., Shigeno C., Atsumi T., Ishizeki K., Suzuki F., Hiraki Y. Chondrogenic differentiation of clonal mouse embryonic cell line ATDC5 in vitro: Differentiation-dependent gene expression of parathyroid hormone (PTH)/PTH-related peptide receptor. J. Cell Biol. 1996;133:457–468. doi: 10.1083/jcb.133.2.457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mortier G.R., Cohn D.H., Cormier-Daire V., Hall C., Krakow D., Mundlos S., Nishimura G., Robertson S., Sangiorgi L., Savarirayan R., et al. Nosology and classification of genetic skeletal disorders: 2019 revision. Am. J. Med. Genet. 2019;179:2393–2419. doi: 10.1002/ajmg.a.61366. [DOI] [PubMed] [Google Scholar]
- 32.Cheng Y., Patel D.J. Crystallographic structure of the nuclease domain of 3′hExo, a DEDDh family member, bound to rAMP. J. Mol. Biol. 2004;343:305–312. doi: 10.1016/j.jmb.2004.08.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thomas M.F., L’Etoile N.D., Ansel K.M. Eri1: A conserved enzyme at the crossroads of multiple RNA-processing pathways. Trends Genet. 2014;30:298–307. doi: 10.1016/j.tig.2014.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.G Walleshauser III J., Kessler T., Morse D., A Tannous B., H L Chiu N., Chiu H.L. A Simple Approach for Evaluating Total MicroRNA Extraction from Mouse Brain Tissues. J. Anal. Sci. Methods Instrum. 2012;02:5–12. [Google Scholar]
- 35.Cox T.C., Lidral A.C., McCoy J.C., Liu H., Cox L.L., Zhu Y., Anderson R.D., Moreno Uribe L.M., Anand D., Deng M., et al. Mutations in GDF11 and the extracellular antagonist, Follistatin, as a likely cause of Mendelian forms of orofacial clefting in humans. Hum. Mutat. 2019;40:1813–1825. doi: 10.1002/humu.23793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gedeon Á.K., Colley A., Jamieson R., Thompson E.M., Rogers J., Sillence D., Tiller G.E., Mulley J.C., Gécz J. Identification of the gene (SEDL) causing X-linked spondyloepiphyseal dysplasia tarda. Nat. Genet. 1999;22:400–404. doi: 10.1038/11976. [DOI] [PubMed] [Google Scholar]
- 37.Bamshad M., Lin R.C., Law D.J., Watkins W.C., Krakowiak P.A., Moore M.E., Franceschini P., Lala R., Holmes L.B., Gebuhr T.C., et al. Mutations in human TBX3 alter limb, apocrine and genital development in ulnar-mammary syndrome. Nat. Genet. 1997;16:311–315. doi: 10.1038/ng0797-311. [DOI] [PubMed] [Google Scholar]
- 38.Lv D., Luo Y., Yang W., Cao L., Wen Y., Zhao X., Sun M., Lo W.H.Y., Zhang X. A novel single-base deletion in ROR2 causes atypical brachydactyly type B1 with cutaneous syndactyly in a large Chinese family. J. Hum. Genet. 2009;54:422–425. doi: 10.1038/jhg.2009.48. [DOI] [PubMed] [Google Scholar]
- 39.De Mori R., Romani M., D’Arrigo S., Zaki M.S., Lorefice E., Tardivo S., Biagini T., Stanley V., Musaev D., Fluss J., et al. Hypomorphic Recessive Variants in SUFU Impair the Sonic Hedgehog Pathway and Cause Joubert Syndrome with Cranio-facial and Skeletal Defects. Am. J. Hum. Genet. 2017;101:552–563. doi: 10.1016/j.ajhg.2017.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bakrania P., Efthymiou M., Klein J.C., Salt A., Bunyan D.J., Wyatt A., Ponting C.P., Martin A., Williams S., Lindley V., et al. Mutations in BMP4 cause eye, brain, and digit developmental anomalies: overlap between the BMP4 and hedgehog signaling pathways. Am. J. Hum. Genet. 2008;82:304–319. doi: 10.1016/j.ajhg.2007.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Elliott A.M., Evans J.A. Genotype-phenotype correlations in mapped split hand foot malformation (SHFM) patients. Am. J. Med. Genet. A. 2006;140:1419–1427. doi: 10.1002/ajmg.a.31244. [DOI] [PubMed] [Google Scholar]
- 42.Chen P., Tan Z., Shek H.T., Zhang J.N., Zhou Y., Yin S., Dong Z., Xu J., Qiu A., Dong L., et al. Phenotypic Spectrum and Molecular Basis in a Chinese Cohort of Osteogenesis Imperfecta With Mutations in Type I Collagen. Front. Genet. 2022;13:816078. doi: 10.3389/fgene.2022.816078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Alexiadis A., Delidakis C., Kalantidis K. Snipper, an Eri1 homologue, affects histone mRNA abundance and is crucial for normal Drosophila melanogaster development. FEBS Lett. 2017;591:2106–2120. doi: 10.1002/1873-3468.12719. [DOI] [PubMed] [Google Scholar]
- 44.Bohnsack K.E., Bohnsack M.T. Uncovering the assembly pathway of human ribosomes and its emerging links to disease. EMBO J. 2019;38:e100278. doi: 10.15252/embj.2018100278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ridanpää M., Van Eenennaam H., Pelin K., Chadwick R., Johnson C., Yuan B., VanVenrooij W., Pruijn G., Salmela R., Rockas S., et al. Mutations in the RNA component of RNase MRP cause a pleiotropic human disease, cartilage-hair hypoplasia. Cell. 2001;104:195–203. doi: 10.1016/s0092-8674(01)00205-7. [DOI] [PubMed] [Google Scholar]
- 46.Vega H., Waisfisz Q., Gordillo M., Sakai N., Yanagihara I., Yamada M., Van Gosliga D., Kayserili H., Xu C., Ozono K., et al. Roberts syndrome is caused by mutations in ESCO2, a human homolog of yeast ECO1 that is essential for the establishment of sister chromatid cohesion. Nat. Genet. 2005;37:468–470. doi: 10.1038/ng1548. [DOI] [PubMed] [Google Scholar]
- 47.Boocock G.R.B., Morrison J.A., Popovic M., Richards N., Ellis L., Durie P.R., Rommens J.M. Mutations in SBDS are associated with Shwachman-Diamond syndrome. Nat. Genet. 2003;33:97–101. doi: 10.1038/ng1062. [DOI] [PubMed] [Google Scholar]
- 48.Ng S.B., Buckingham K.J., Lee C., Bigham A.W., Tabor H.K., Dent K.M., Huff C.D., Shannon P.T., Jabs E.W., Nickerson D.A., et al. Exome sequencing identifies the cause of a mendelian disorder. Nat. Genet. 2010;42:30–35. doi: 10.1038/ng.499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dixon M.J. Treacher Collins syndrome. Hum. Mol. Genet. 1996;5:1391–1396. doi: 10.1093/hmg/5.supplement_1.1391. [DOI] [PubMed] [Google Scholar]
- 50.Farley-Barnes K.I., Ogawa L.M., Baserga S.J. Ribosomopathies: Old Concepts, New Controversies. Trends Genet. 2019;35:754–767. doi: 10.1016/j.tig.2019.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Carapito R., Konantz M., Paillard C., Miao Z., Pichot A., Leduc M.S., Yang Y., Bergstrom K.L., Mahoney D.H., Shardy D.L., et al. Mutations in signal recognition particle SRP54 cause syndromic neutropenia with Shwachman-Diamond-like features. J. Clin. Invest. 2017;127:4090–4103. doi: 10.1172/JCI92876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bellanné-Chantelot C., Schmaltz-Panneau B., Marty C., Fenneteau O., Callebaut I., Clauin S., Docet A., Damaj G.L., Leblanc T., Pellier I., et al. Mutations in the SRP54 gene cause severe congenital neutropenia as well as Shwachman-Diamond – Like syndrome. Blood. 2018;132:1318–1331. doi: 10.1182/blood-2017-12-820308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Trainor P.A., Merrill A.E. Ribosome biogenesis in skeletal development and the pathogenesis of skeletal disorders. Biochim. Biophys. Acta. 2014;1842:769–778. doi: 10.1016/j.bbadis.2013.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Han M., Chang M., Kim U.J., Grunstein M. Histone H2B repression causes cell-cycle-specific arrest in yeast: Effects on chromosomal segregation, replication, and transcription. Cell. 1987;48:589–597. doi: 10.1016/0092-8674(87)90237-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The RNA-seq data have been deposited in NCBI GEO with the accession code GEO: GSE206412. The individuals’ genomic data obtained by exome or genome sequencing are not publicly available because study participants did not give full consent for releasing data publicly. These data can be accessed under the condition that a joint research plan is made by the researchers and approved by the ethics committees of the participating institutions. All the other data supporting the findings of this study are included within the article or the supplemental information.