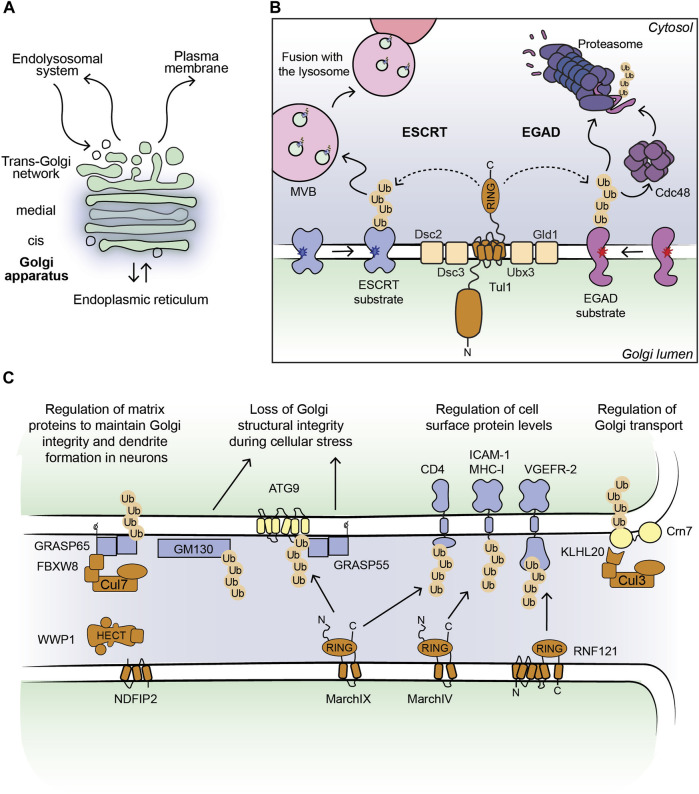
FIGURE 1.
(A) Outline of a Golgi stack. (B) Cartoon model of the Dsc complex in yeast targeting substrates for ESCRT- and EGAD-mediated degradation. Non-native proteins recognized by the Dsc complex are shown in blue (ESCRT substrates) and purple (EGAD substrates). Dark blue and red stars indicate folding lesions within the TM domains of the substrate proteins. The Tul1 ubiquitinated ESCRT substrates are sorted into ILVs by the ESCRT machinery (not shown for simplicity). MVBs containing the ILVs fuse with the lysosome/vacuole and ubiquitinated proteins are degraded. The Tul1 ubiquitinated EGAD substrates are removed from the Golgi membrane and unfolded by Cdc48, and eventually degraded by the proteasome. (C) Current outline of E3 ligase-substrate pairs at the Golgi apparatus in mammalian cells. Selected E3 ligases (shown in brown) with their known substrates, where the ubiquitinated proteins can be targeted for degradation (shown in blue) or induce changes in protein-protein interactions (shown in yellow). The consequences of protein ubiquitination at the Golgi are stated in the text above.