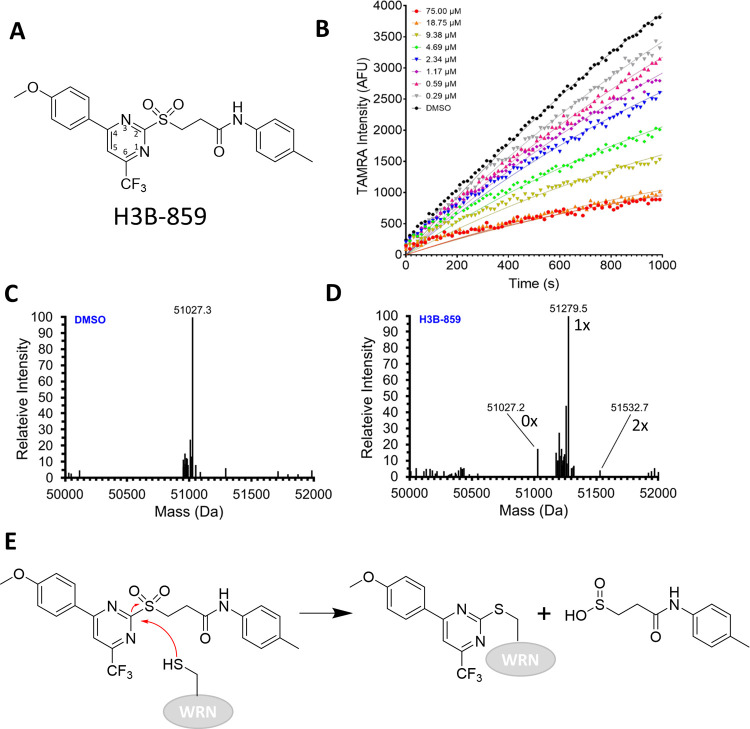
Figure 2.
Evidence for covalent modification of WRN by H3B-859. (A) Chemical structure for starting lead H3B-859. The pyrimidine ring is numbered according to convention. (B) Real-time DNA unwinding progress curves with 0.6 nM WRN helicase domain, 0.5 μM Hel-10bp, 5 μM Trap-10bp, 120 μM ATP, and the indicated concentrations of H3B-859. (C) Intact mass analysis of WRN helicase domain (expected MW = 51026.6 Da) with DMSO. (D) Intact mass analysis after overnight treatment with a 10-fold excess of H3B-859. Alkylation by H3B-859 is expected to shift the mass by 253 Da. Masses corresponding to unmodified, monoalkylated, and dialkylated proteins are indicated in the spectrum as 0×, 1×, and 2×, respectively. (E) Proposed mechanism of inhibition by 2-sulfonylpyrimidine. A cysteine from WRN undergoes nucleophilic aromatic substitution to form the indicated adduct with departure of the sulfinic acid leaving group.