Figure 3.
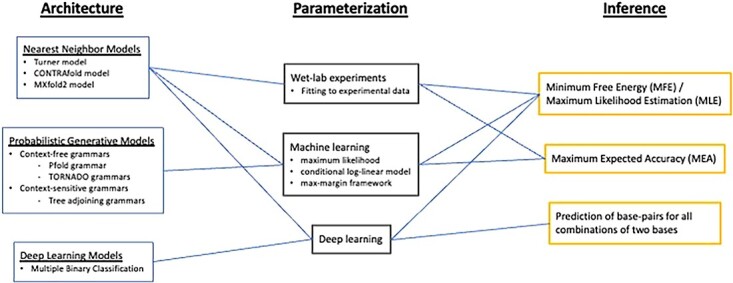
A schematic diagram of de novo RNA secondary structure prediction algorithms. Most RNA secondary structure prediction algorithms can be categorized by three aspects: ‘Architecture’, ‘Parameterization’ and ‘Inference’. Rivas et al. [25] added ‘Scoring scheme’ to these aspects, which is uniquely determined by ‘Parameterization’, and thus ‘Scoring scheme’ is omitted in this paper. From the ‘Architecture’ aspect, RNA secondary structure prediction algorithms can be categorized into nearest neighbor models, probabilistic generative models and deep learning models, depending on the RNA computational models. These are further sub-categorized according to their parameter assignment, fine-grainedness, grammatical rules etc. The ‘Parameterization’ aspect classifies RNA secondary structure prediction algorithms into three types, depending on how they find optimal parameter values for the parameter set defined in the ‘Architecture’: wet-lab experiments, machine learning and deep learning. Finally, the ‘Inference’ aspect classifies RNA secondary structure prediction algorithms according to how they use the models determined in the ‘Archtecture’ and ‘Parameterization’ to make secondary structure predictions.
