Table 1.
List of de novo RNA secondary structure prediction tools presented in this paper that are currently available.
The column labeled ‘Model’ indicates the category to which the method belongs (NN: nearest neighbor model, PG: probabilistic generative model, DL: deep learning-based model). The ‘TM’ column indicates whether the method uses thermodynamic parameters. The column labeled ‘ML’ indicates whether the method can train its parameters using machine learning, and if so, which training method is used (MLE: maximum likelihood estimation, CG: constraint generation, CLLM: conditional log-linear models, BL: Boltzmann likelihood, MM: max-margin framework, DL: deep larning). The ‘MEA’ column indicates whether the method predicts secondary structure by default (✓) or optionally (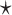 ) with the maximum expected accuracy. The column labeled ‘PK’ indicates whether the method can predict pseudoknotted structures.
) with the maximum expected accuracy. The column labeled ‘PK’ indicates whether the method can predict pseudoknotted structures.