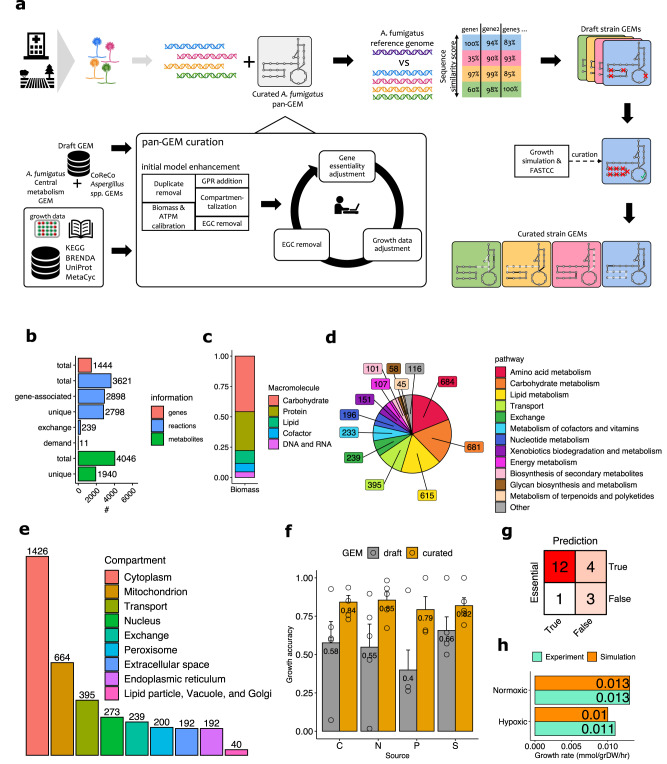
Fig. 1. General reconstruction workflow and A. fumigatus pan-genome-scale metabolic model (GEM) statistics.
a Workflow for A. fumigatus strain-specific GEM reconstructions. Colors indicate strains and associated metabolic models. b–g Characteristics of pan-GEM reconstruction for A. fumigatus. b Counts of pan-GEM components for included genes, reactions, and metabolites. c Contribution of macromolecules in one unit of biomass (Supplementary Data S1). d Distribution of pan-GEM reactions across major pathway categories (Supplementary Data S9). e Distribution of pan-GEM reactions across nine compartments (Supplementary Data S9). f Growth prediction accuracy of pan-GEM for A. fumigatus wild-type (Af293) and four mutant strains using phenotypic microarray data (n = 5 in total, bars show mean and standard error of mean, Supplementary Data S2). C: carbon, N: nitrogen, P: phosphorus, S: sulfur. g Confusion matrix of pan-GEM accuracy in predicting the essentiality of 20 genes according to the literature (see Results and Methods). h Experimental values compared to simulated growth rate values under normoxic and hypoxic conditions (Supplementary Data S2 has experimental and simulated secretion values). Source data for Fig. 1b–h are provided in the Source Data file.