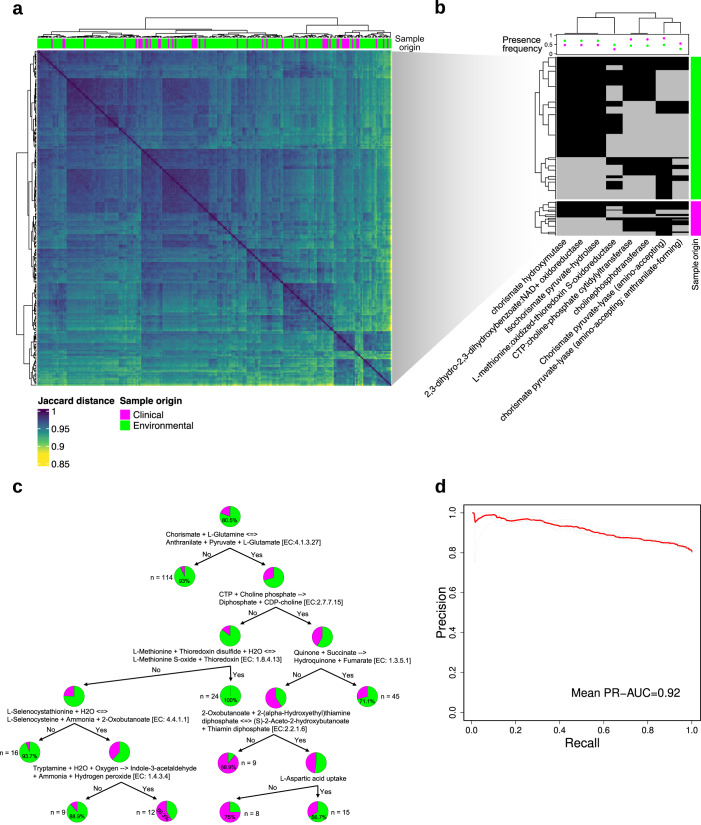
Fig. 3. Differentiation by origin of all strain-specific GEMs.
a Heatmap with pairwise Jaccard distance values for isolated GEM pairs based on presence or absence of metabolic reactions. b Selected metabolic reactions with the highest statistical significance for differences in presence/absence by sample origin (Fisher exact test, p ≤ 0.05). Presence frequency indicates the fraction of reaction presence over all investigated strain-specific GEMs. c Decision tree optimized for separation of clinical and environmental strain origin. The decision tree is based on absence or presence of metabolic reactions and growth capability on different nutrients for all strain-specific GEMs. d Machine learning-derived mean area under the curve for precision over recall based on 21 reactions identified by the model using biomass-supporting flux ranges for all reactions determined with flux variability analysis to classify clinical vs. environmental origin. Source data for Fig. 3 are provided in the Source Data file.