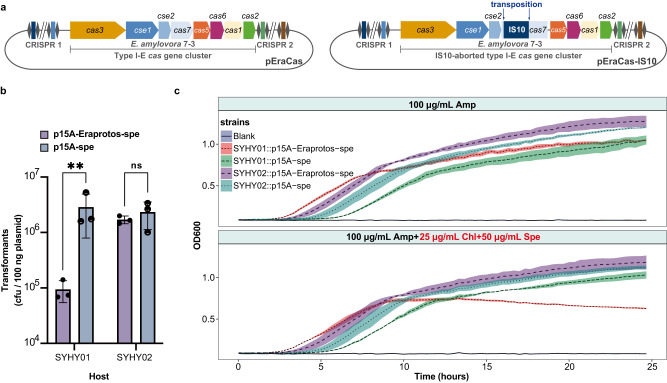
Fig. 2. ISs-deactivated CRISPR-Cas system facilitates the rapid acquisition and improved persistence of the antibiotic resistance plasmid containing identifiable protospacers.
a Schematic of the engineered type I-E CRISPR-Cas locus of E. amylovora 7-3. An intact or IS10-aborted CRISPR-Cas locus was cloned into plasmid pEraCas or pEraCas-IS10, respectively, for CRISPR-Cas interference assays. b Plasmid transformation interference by the E. amylovora 7-3 type I-E CRISPR-Cas system in E. coli DH10B cells. Transformation efficiency was determined by counting colony-forming units (CFUs) per 100 ng of plasmid. Data shown as means ± S.D. from three biological replicates; **P < 0.01; ns not significant, using two-tailed unpaired t-test of log10 transformed data; P = 0.0018, 0.6150 (from left to right). Source data are provided as a Source Data file. c Growth curves of test strains under different antibiotic exposures. Shaded areas indicate the means ± S.D. from three biological replicates. Ampicillin (Amp) was used to maintain pEraCas or pEraCas-IS10 in the host, and chloramphenicol (Chl) and spectinomycin (Spe) were used to maintain p15A-Eraprotos-spe and p15A-spe. Source data are provided as a Source Data file.