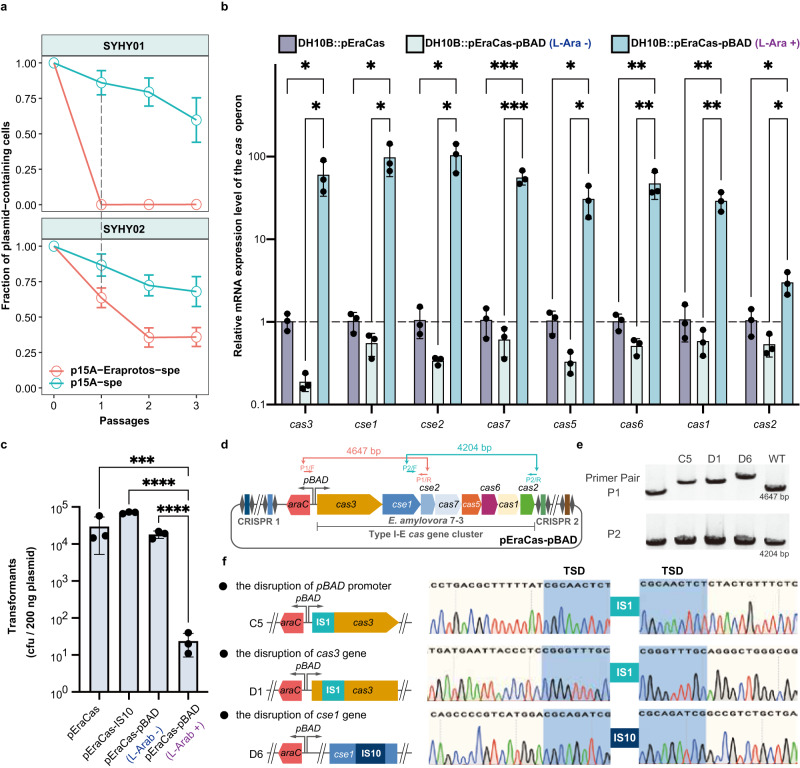
Fig. 3. ISs mediate the fitness trade-off between the benefits of acquiring plasmids and genetic denfense under selective pressure.
a Plasmid stability after transformation and during passage in SYHY01 and SYHY02. Values shown as mean ± SEM from three biological replicates. Source data are provided as a Source Data file. b Relative transcription levels of eight genes within the cas operon of pEraCas and pEraCas-pBAD in E. coli DH10B. Gene expression was quantified by RT-qPCR and normalized to levels of endogenous 16sRNA. Error bars denote means ± S.D. from three biological replicates; *P < 0.05; **P < 0.01; ***P < 0.001, using two-tailed unpaired t-test; all the presented P-values were displayed from top to bottom, 0.019, 0.019 (cas3); 0.015, 0.014 (cse1); 0.011, 0.011 (cse2); 0.000886, 0.000858 (cas7); 0.017, 0.016 (cas5); 0.009, 0.009 (cas6); 0.003, 0.003 (cas1); 0.028, 0.011 (cas2). Source data are provided as a Source Data file. c Transformation efficiencies of pEraCas (targeting the SYHY03 chromosome), pEraCas-IS10 (control), and pEraCas-pBAD (targeting the SYHY03 chromosome in the presence of l-arabinose) into SYHY03 were determined through counting CFUs per 200 ng of plasmids. Error bars denote means ± S.D. from three biological replicates; ***P < 0.001; ****P < 0.0001, using two-tailed unpaired t-test of log10 transformed data. P = 0.000256, 2.66e-05, 7e-05 (from top to bottom). Source data are provided as a Source Data file. d Schematic of pEraCas-pBAD harboring the L-arabinose-inducible pBAD promoter to drive cas operon expression. Primer pairs P1 and P2 were designed to amplify the entire cas operon of pEraCas-pBAD; expected amplicon sizes are shown. e Colony PCR screening with primer pairs P1 and P2 for IS insertions into the pEraCas-pBAD cas operon. Lanes “WT”, control amplicons with the pEraCas-pBAD template; larger PCR products (lanes C5, D1, and D6) suggest IS transposition events. The experiment was conducted independently three times and yielded consistent results. Source data are provided as a Source Data file. f Confirmation by Sanger sequencing of IS insertions into cas operons of mutants C5, D1, and D6. Shaded areas of chromatograms, TSDs resulting from IS insertions.