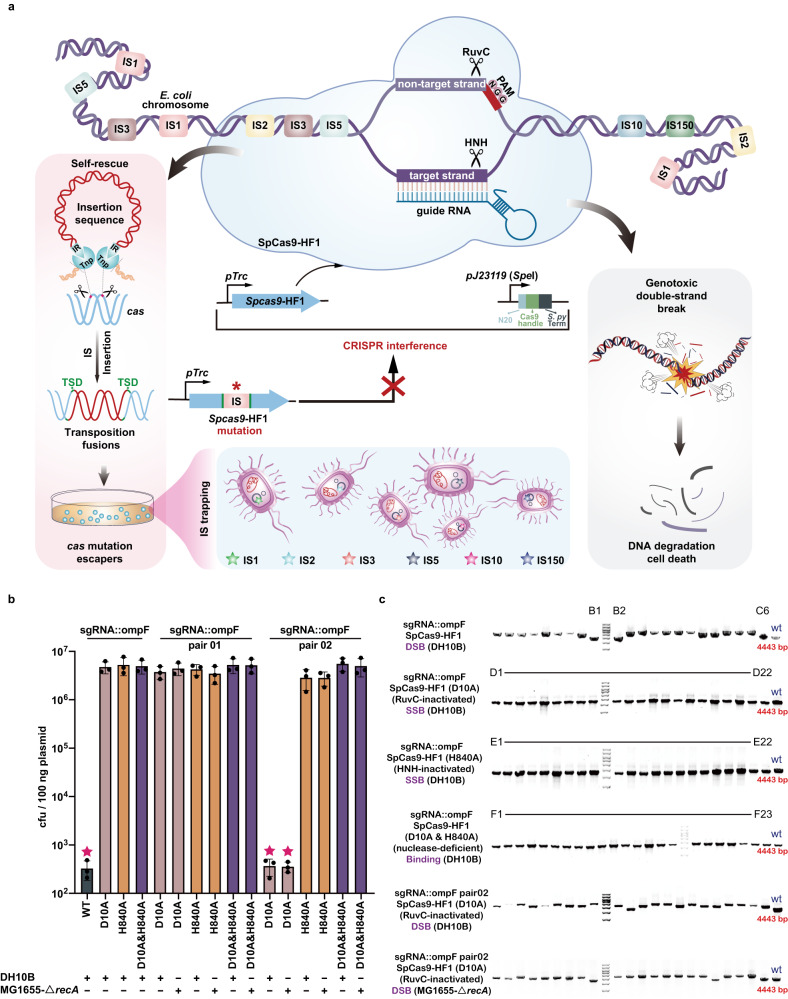
Fig. 4. Establishment and characterization of the CRISPR/Cas-mediated IS trapping system.
a Schematic of the IS-trapping system in which DSBs trigger transposition of IS elements, including insertions into cas. b Transformation efficiency of the sgRNA-expressing interference plasmid in E. coli DH10B and MG1655-ΔrecA harboring CRISPR-Cas (WT) or CRISPR-Cas mutants. Mutants were generated by site-directed mutagenesis in D10A and H840A within the SpCas9-HF1 coding region, inactivating the RuvC and HNH domains, respectively. sgRNA::ompF, sgRNA::ompF pair 01, and sgRNA::ompF pair 02 represent targeting plasmids p15A-cm-sgRNA::ompF, p15A-cm-sgRNA::ompF pair 01, and p15A-cm-sgRNA::ompF pair 02, respectively. The five-pointed star above bars indicates the strain has a CRISPR/Cas-induced double-strand break in the chromosome. The “+” or “−” indicate that the given E. coli strain was used or not used in the corresponding sample in the bar graph. WT, D10A, H840A, and D10A&H840A indicate plasmids pCasY-3-ΔSIM-kan, pCasY-3-ΔSIM-kan (D10A), pCasY-3-ΔSIM-kan (H840A), and pCasY-3-ΔSIM-kan (D10A&H840A), respectively; error bars indicate means ± S.D. from three biological replicates. Source data are provided as a Source Data file. c PCR screening for IS insertions into cas gene expression cassettes. The single colonies were taken from each of the four types of CRISPR-tolerant mutants from Fig. 4b, simultaneously containing the genomic target sequence, sgRNA expression plasmid, and CRISPR-Cas expression cassette [pTrc-SpCas9-HF1, pTrc-SpCas9-HF1 (D10A), pTrc-SpCas9-HF1 (H840A), or pTrc-SpCas9-HF1 (D10A&H840A)], followed by colony PCR validation. The expected size of PCR amplicons from the four types of cas gene expression cassettes without IS element insertions was 4443 bp; larger PCR products indicate IS insertion. DSB denotes double-stranded DNA breaks, while SSB represents single-stranded DNA breaks. Binding refers to the specific binding of Cas protein to the target site without inducing DNA cleavage. The experiment was conducted independently three times and yielded consistent results. Source data are provided as a Source Data file.