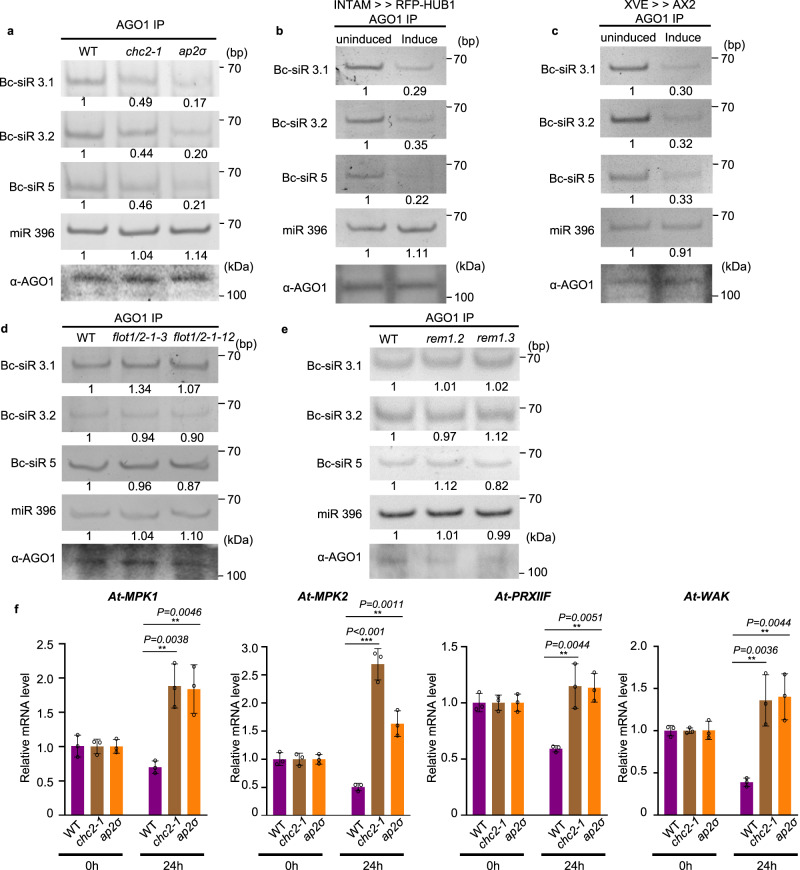
Fig. 4. Bc-sRNA loading into plant AGO1 and target gene suppression was reduced in CME mutants.
a The loading of Bc-siR3.1, Bc-siR3.2, and Bc-siR5 into AGO1 of wild-type, chc2-1, and ap2σ mutants was examined with AGO1-IP followed by RT-PCR. b, c, The loading of Bc-siR3.1, Bc-siR3.2, and Bc-siR5 into AGO1 of induced INTAM≫RFP-HUB1 (b) and XVE≫AX2 (c) transgenic plants was examined with AGO1-IP followed by RT-PCR. d, e The loading of Bc-siR3.1, Bc-siR3.2, and Bc-siR5 into AGO1 of wild-type, flot1/2 (d), rem1.2, and rem1.3 mutants (e) was examined with AGO1-IP followed by RT-PCR. In a–e, miR396 served as a positive control. AGO1 was detected by western blot. f, Suppression of host target genes by Bc-sRNAs was attenuated in CME mutants. The expression of Bc-siR3.1 target At-PRXIIF, Bc-siR3.2 target At-MPK1 and At-MPK2, and Bc-siR5 target At-WAK in chc2-1 and ap2σ mutants compared with those in wild-type infected plants after B. cinerea infection was examined by real-time RT-PCR. The data are presented as mean ± s.d. Similar results were obtained in three biologically independent experiments. The statistical analysis was performed using ANOVA Dunnett’s multiple comparisons test. The small open circles represent individual values. The error bars indicate s.d. *P < 0.05, **P < 0.01, ***P < 0.001. Source data are provided as a Source Data file.