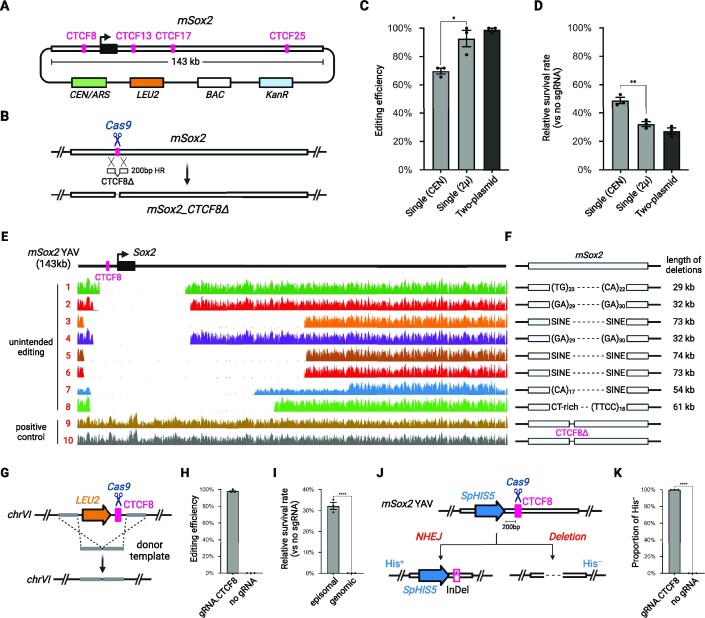
Figure 3.
Episomal editing in yeast targeting at mSox2 YAV. (A) Structure of episomal mSox2 YAV. BAC, bacteria artificial chromosome backbone. (B) Episomal editing strategy. Cas9 creates a DSB at CTCF8 which can be repaired with a donor DNA containing CTCF8Δ. (C) Editing efficiency with the single-plasmid (CEN or 2μ backbone) or two-plasmid (CEN/2μ) systems, as determined by genotyping using deletion-specific primers (Supplementary Figure S1). Error bars represent mean ± SD of three replicates. P< 0.05 (*), as calculated by an unpaired t-test. (D) Survival rate of colonies transformed with Cas9/gRNA targeting CTCF8 without donor DNA templates. Error bars represent mean ± SD of three replicates. P< 0.005 (**), as calculated by an unpaired t-test. (E) WGS read coverage of colonies with failed or successful edits aligned to the mSox2 YAV. (F) Unintended deletion boundaries mapped to the indicated mouse genome repetitive elements (see also Supplementary Table S6). (G) Editing strategy of a genome-integrated mSox2 CTCF8 site inserted between YFL021C and YFL021W. (H) Editing efficiency of a genome-integrated mSox2 CTCF8. Error bars represent mean ± SD of three replicates (n = 32 for each group). (I) Survival rate of colonies for genomic and episomal YAV editing without donor DNA templates. Error bars represent mean ± SD of three replicates. P< 0.0001 (****), as calculated by an unpaired t-test. (J) Episomal YAV editing with an SpHIS5 marker adjacent to CTCF8 site. His+ or His–, prototrophic or auxotrophic for histidine, respectively. (K) Ratio of His– colonies surviving episomal YAV editing (shown in J). Error bars represent mean ± SD of three replicates. P< 0.0001 (****), as calculated by an unpaired t-test. Representative images of transformation plates are depicted in Supplementary Figure S5).