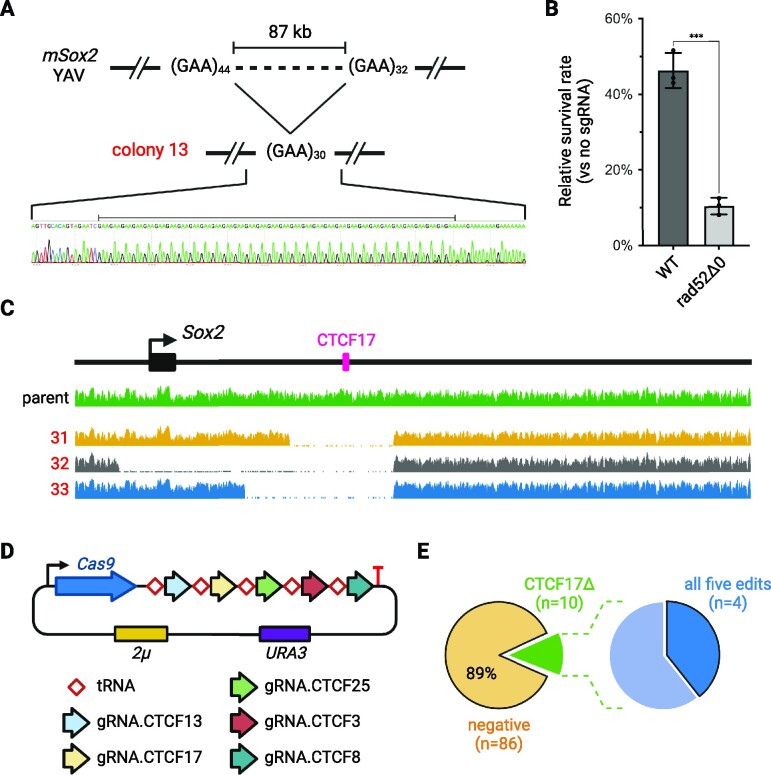
Figure 5.
Internal deletion mechanism and multiplex editing with five targets. (A) Junctions from the internal deletions in colony 13. More junction sequences are shown in Supplementary Figure S9. (B) Survival rate with Cas9 and gRNA.CTCF17, in wild-type or rad52Δ0 background. (C) WGS read coverage from three colonies obtained in CTCF17 cleavage experiments in rad52Δ0 strain background, aligned to the original mSox2 YAV. (D) CREEPY construct used for multiplex editing with five targets. (E) Editing efficiency with five targets. Colonies (n = 96) were first screened for CTCF17Δ (left). Positive colonies (n = 10, green) were subsequently selected for further screening of all other targets. The raw PCR screening gel image is shown in Supplementary Figure S12.