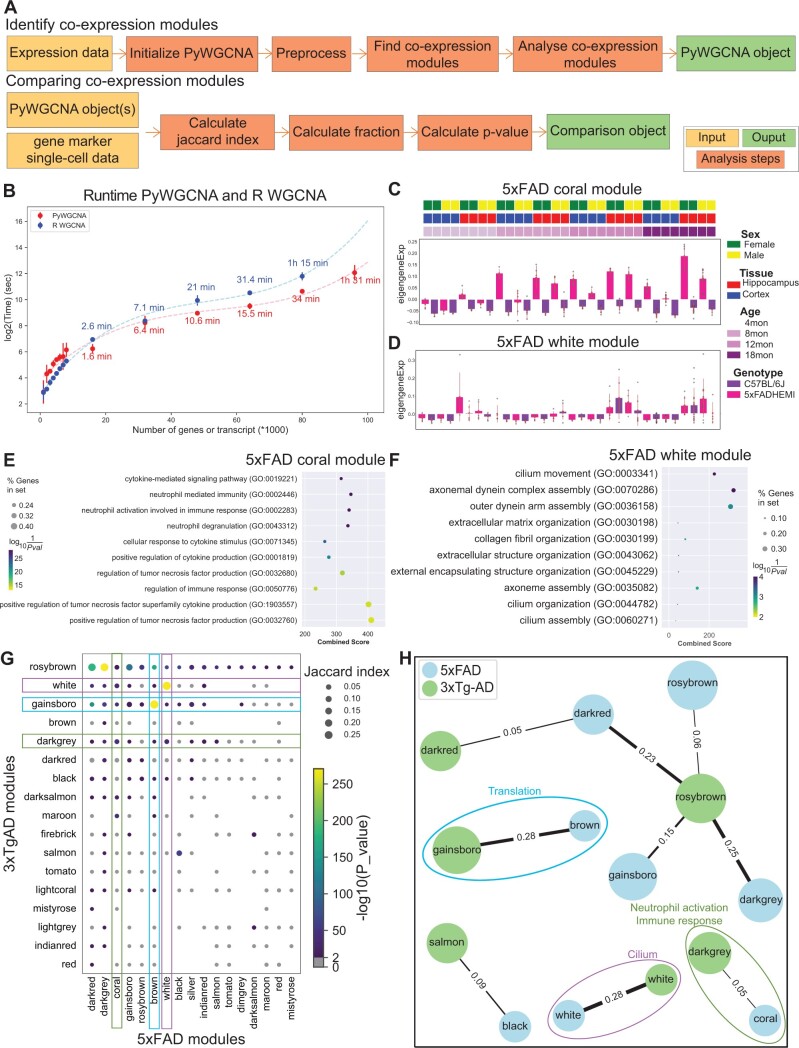
Figure 1.
PyWGCNA steps and output example. (A) Overview of PyWGCNA. (B) Average runtime of PyWGCNA and R version of WGCNA versus the number of features used in downsampled datasets in triplicate. (C) Coral and (D) white module eigengene expression profile from 5xFAD mouse model summarized by genotype. Above, the top three rows display the metadata for each dataset including sex, tissue, and age. Below, the bar plot represents module eigengene expression by genotype for each dataset with individual sample module eigengene expression shown as points. GO analysis of the genes in 5xFAD (E) coral and (F) white modules, respectively. (G) Bubble plot of module overlap test results between 5xFAD and 3xTg-AD mouse models of familial AD. The dot size represents the fraction of shared genes between each pair of modules and non-gray color denotes the significance of the overlap between modules. (H) Comparison graph of 5xFAD and 3xTg-AD modules mouse models of familial AD for those with >0.05 Jaccard similarity. The thickness of lines shows the Jaccard index value.