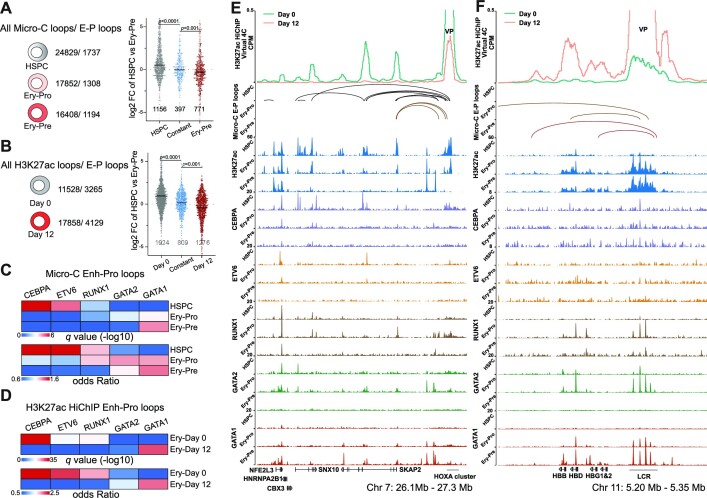
Figure 3.
Stage-specific transcription factors are involved in the rewiring of enhancer–promoter contacts. (A) The number of all loops and E–P loops detected by Micro-C (left) in HSPC, Ery-Pro and Ery-Pre; the gene expression (log2FC HSPC versus Ery-Pre) of corresponding promoter anchors (right) in HSPC specific, constant and Ery-Pre specific loops. Statistical significance was determined using the Mann–Whitney test. (B) The number of all loops and E–P loops detected by H3K27ac-HiChIP (left) in Ery-Day0 and Ery-Day12; the gene expression (log2FC HSPC versus Ery-Pre) of corresponding promoter anchors (right) in HSPC specific, constant and Ery-Pre specific loops. Statistical significance was determined using the Mann–Whitney test. (C) LOLA enrichment analysis for the enhancer anchors of the Micro-C E–P loops in HSPC, Ery-Pro and Ery-Pre stages using our CUT&RUN datasets of transcription factors. LOLA enrichment for each transcription factor was based on combined peaks from all stages of the respective transcription factor. (D) LOLA enrichment analysis for the enhancer anchors of the H3K27ac-HiChIP E–P loops in erythroid culture day 0 (Ery-Day0) and day 12 (Ery-Day12) using our CUT&RUN datasets of transcription factors. LOLA enrichment for each transcription factor was based on combined peaks from all stages of the respective transcription factor. (E) Virtual 4C profile of normalized H3K27ac HiChIP signals around HOXA9 VPs (Chr 7: 26.1–27.3 Mb, top), Micro-C detected E–P loops (middle), and CUT&RUN signals of H3K27ac, GATA1, GATA2, ETV6, CEBPA and RUNX1 (bottom) at HSPC, Ery-Pro and Ery-Pre stages. (F) Virtual 4C profile of normalized H3K27ac HiChIP signals around LCR VPs (Chr 11: 5.20–5.35 Mb, top), Micro-C detected E–P loops (middle), and CUT&RUN signals of H3K27ac, GATA1, GATA2, ETV6, CEBPA and RUNX1 (bottom) at HSPC, Ery-Pro and Ery-Pre stages. Abbreviations: LOLA, locus overlap analysis; CPM, counts per million; VP, viewpoint.