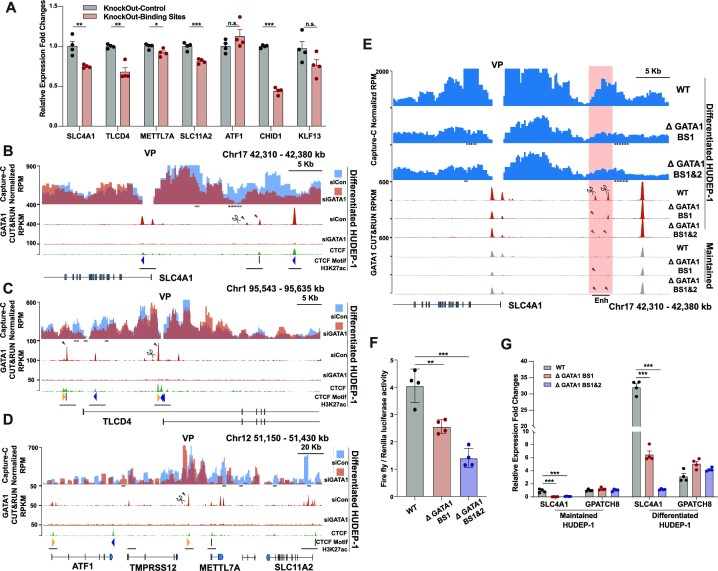
Figure 5.
Ery-Pre cells acquired GATA1 occupancy sites that promote gene expression and local chromatin rewiring. (A) Gene expression of SLC4A1, TLCD4, METTL7A, ATF1, SLC11A2, CHID1 and KLF13 after knock-out of the corresponding gained GATA1 binding sites in Ery-Pre cells (see panel 5B–D and Figure 4) in differentiated HUDEP-1 cells. Data are shown as mean ± SEM (n = 4). Statistical significance was determined using the Student's t-test; *P < 0.05, **P < 0.01, ***P < 0.001. (B–D) Top, Capture-C contact profiles plotted as overlays, comparing contacts of the control (siCon, blue) and GATA1 knock-down (siGATA1, red) in differentiated HUDEP-1 cells at the SLC4A1 locus (Chr17: 42310–42380 kb) (B), TLCD4 locus (Chr1: 95 543–95 653 kb) (C) and METTL7A locus (Chr12: 51 150–51 430 kb) (D). The y-axis shows the Capture-C sequencing coverage expressed as mean RPM with 200 bp bin and 2000 bp window (B, C) or 200 bp bin and 4000 bp window (D) (n = 2). VP represents the viewpoint of the SLC4A1 promoter (B), TLCD4 intronic GATA1 binding site (C) and METTL7A distal GATA1 binding site (D). Bottom, GATA1 CUT&RUN tracks of siCon and siGATA1, respectively. Arrowheads indicate the gained GATA1 peaks in the erythroid progenitor-precursor transition. CTCF CUT&RUN signal, CTCF-site orientations, and H3K27ac peaks are indicated at the bottom. Dash lines below the Capture-C contact profiles indicate statistically significant regions (EDGER, P < 0.05, log2FC > 1.5). (E) Top, Capture-C contact profiles plot, comparing contacts of the WT, single GATA1 binding site mutant (ΔGATA1 BS1), and dual GATA1 binding site mutant (ΔGATA1 BS1&2) in differentiated HUDEP-1 clones at the SLC4A1 locus (Chr17: 42 310–42 380 kb). The y-axis shows Capture-C sequencing coverage expressed as mean RPM with 200 bp bin and 2000 bp window (n = 2). VP represents the viewpoint of the SLC4A1 promoter. Bottom, GATA1 CUT&RUN tracks of WT, single GATA1 binding site mutant (ΔGATA1 BS1), and dual GATA1 binding site mutant (ΔGATA1 BS1&2) HUDEP-1 clones in differentiated and maintained stages, respectively. Arrowheads show the degenerated peaks. Enh indicates the enhancer sequence used for the reporter gene assay in panel F (Chr17: 42 358 715–42 361 352 bp). Dash lines below the Capture-C contact profiles indicate statistically significant regions (EDGER, P < 0.05, log2FC > 1.5). (F) Enhancer reporter gene assay of WT, ΔGATA1 BS1, and ΔGATA1 BS1&2 enhancer with the SLC4A1 promoter (mean ± SEM, n = 4). Statistical significance was determined using the Student's t-test, ** P < 0.01, *** P < 0.001. (G) Gene expression of WT, ΔGATA1 BS1, and ΔGATA1 BS1&2 HUDEP-1 clones in undifferentiated (maintained) and differentiated condition (mean ± SEM, n = 4, normalized to maintained WT). Statistical significance was determined using the Student's t-test; ** P < 0.01, *** P < 0.001. GPATCH8 is the reference gene located distal to SLC4A1 but not controlled by GATA1. Abbreviations: WT, wild-type; RPM, reads per million; VP, viewpoint