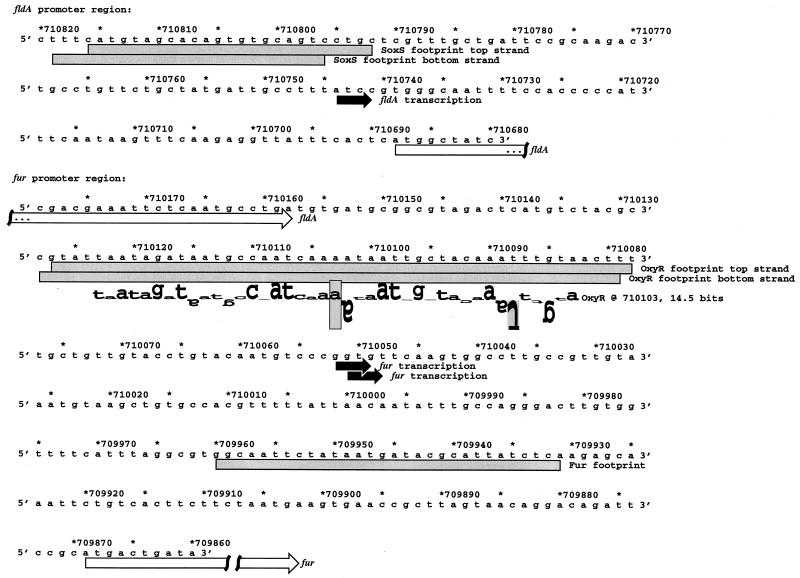
FIG. 1.
Sequences of fur and fldA promoter regions. The DNA sequences and coordinates are for E. coli from GenBank accession no. U00096 (5). The transcriptional starts are marked by black arrows, the fldA and fur open reading frames are indicated by white arrows, and the DNase I footprints are denoted by gray boxes. The location of the predicted OxyR site is shown by a sequence walker (27), in which the rectangle surrounding the adenine at position 710103 indicates the center (zero base) of the binding site. A sequence walker consists of a string of letters in which the height of each letter shows the contribution that the corresponding base would make to the average sequence conservation shown by the sequence logo of all binding sites (25). The sequence walker is given on a scale of bits of information. The rectangle surrounding the adenine gives the scale, from −3 bits up to the maximum conservation at +2 bits. Positively contributing bases are above the zero line, while negatively contributing ones are below the line. By using bits, the heights of all letters can be added together to obtain the total sequence conservation of a site. The fur site is 14.5 bits, and the average OxyR site is 16.7 ± 1.9 bits. The rectangle surrounding the inverted thymine at position 710088 indicates that no T residues were found at this position in the OxyR sites used to construct the model.