FIGURE 7.
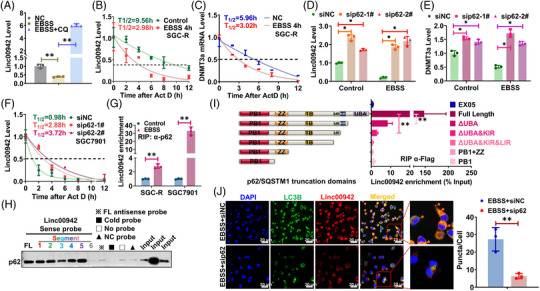
p62 mediated RNautophagic degradation of Linc00942. (A) The effect of autophagy induction or inhibition on Linc00942 level in SGC7901 is determined by qRT‐PCR. (B) The half‐life of Linc00942 in SGC‐R cells before and after EBSS treatment was determined by qRT‐PCR. Half‐life curves were plotted on non‐linear fitting and regression (one‐phase decay curve fit). (C) The half‐life of DNMT3a mRNA in SGC‐R cells treated as indicated was determined by RT‐PCR. (D) The effect of p62 KD on Linc00942 expression before and after EBSS treatment is determined by qRT‐PCR. (E) The effect of p62 KD on DNMT3a mRNA before and after EBSS treatment is determined by qRT‐PCR. (F) The half‐life of Linc00942 in SGC7901 cells before and after p62 KD was determined by RT‐PCR. (G) The effect of autophagy activation on the interaction of p62 with Linc00942 was analysed by RIP assay. (H) The interaction of p62 with various Linc00942 segments was analysed by RNA pull‐down assay. (I) The interaction of Linc00942 with various p62 truncations was analysed by RIP assay. (J) The colocalization of Linc00942 with LC3B in SGC‐R cells treated as indicated was analysed by combined FISH and IF assay (original magnification, 100X). Scale bar: 20 μm.
