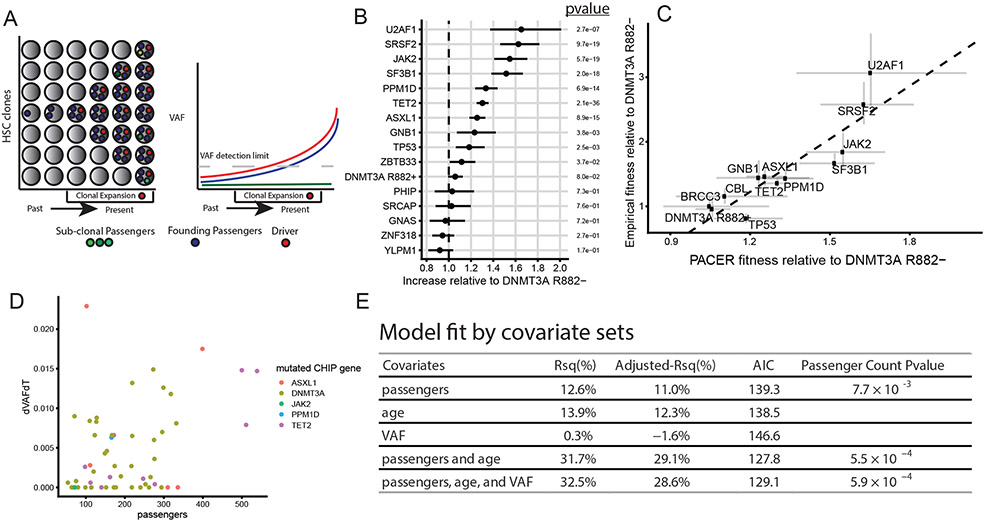
Fig 1∣. PACER Enables Estimation of Clonal Expansion Rate from a Single Blood Draw.
A, A schematic depiction of using passenger counts to estimate the rate of expansion of a hematopoietic stem cell (HSC) clone after the acquisition of a driver mutation. The passengers (blue) that precede the driver (red) can be used to date the acquisition of the driver. B, The relative abundances of passenger counts were estimated for CHIP driver genes with at least 30 cases using a negative binomial regression, adjusting for age at blood draw, driver VAF, and study. The total number of CHIP carriers included is 4,536. The coefficients are relative to DNMT3A R882- CHIP. Unadjusted, two-sided p-values are reported. Error bars indicate 95 percent confidence intervals. C, The relative abundances of passenger counts are plotted against the empirical estimates of gene fitness derived from the longitudinal deep sequencing in Fabre et al.16. Error bars indicate 95 percent confidence intervals. The estimate of the association from weighted least squares (slope = 2.7, p-value = 9.6 x 10−5, R2 = 80%) is plotted as a dashed line. D, The observed clonal expansion rates (dVAFdT), as expressed in the change in variant allele frequency (VAF) over time (years), were associated with increased passenger counts in 55 CHIP carriers from the Women’s Health Initiative. Colors indicate the mutated driver gene. E, A multivariable model including passenger counts, age at blood draw, and VAF indicates the relative contributions of age and VAF over baseline models. AIC is Akaike information criteria, where smaller values indicate better model fit. Unadjusted, two-sided p-values are reported for the passengers variable in the respective models.