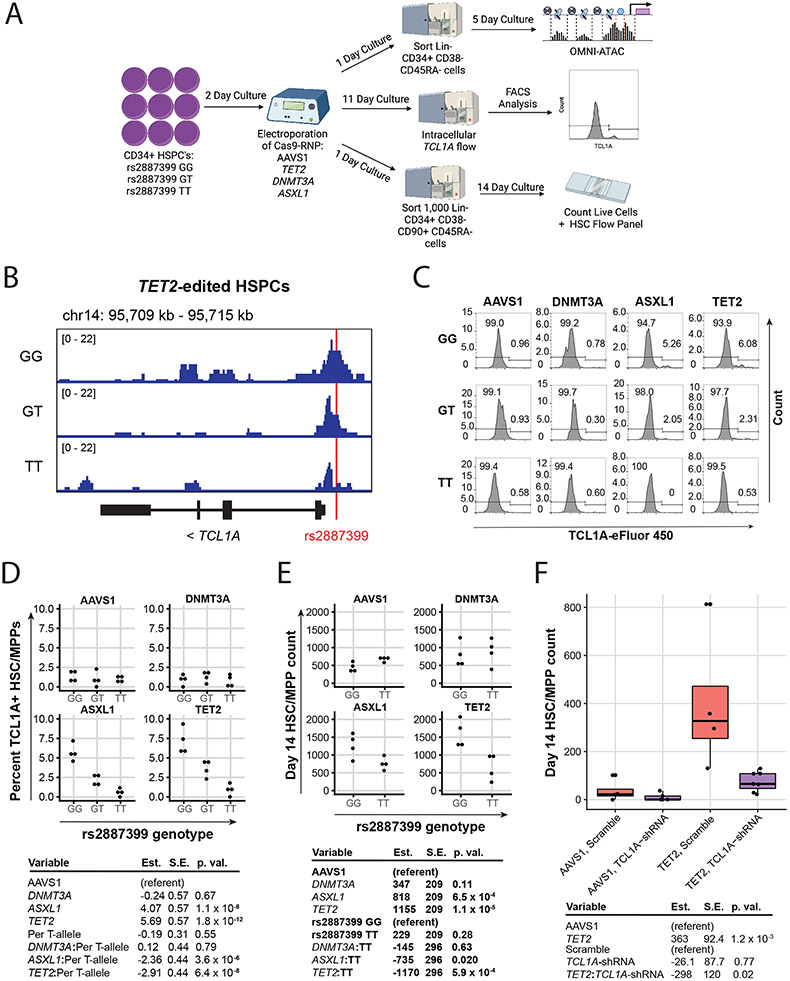
Figure 3∣. Effect of rs2887399 on TCL1A Expression and Clonal Expansion.
A, Schematic of experimental workflow. B, ATAC-sequencing tracks illustrating chromatin accessibility at rs2887399 in TET2-edited HSPCs from donors of the GG, GT, and TT genotypes after 5 days liquid culture. Red line indicates location of rs2887399. See also Extended Data Figure 8 and Table S14. C, Percent Lin− CD34+ CD38− CD45RA− cells expressing TCL1A by flow cytometry after 11 days liquid culture of edited HSPCs, stratified by edited gene and rs2887399 genotype. Results of a linear regression model for the effect of edited gene (referent to AAVS1), number of T-alleles at rs2887399, and the interaction term of edited gene with T-alleles are presented below. Est. = estimate, S.E. = standard error, p. val. = p-value. Unadjusted p-values from a two-sided test are reported. n=4 biologically independent replicates for each group. D, Lin− CD34+ CD38− CD45RA− cell counts after 14 days liquid culture of edited HSCs. Results of a linear regression model for the effect of edited gene (referent to AAVS1), rs2887399 genotype (referent to GG), and the interaction term of edited gene with rs2887399 genotype are presented below. Unadjusted p-values from a two-sided test are reported. n=4 biologically independent replicates for each group. E, Lin− CD34+ CD38− CD45RA− cell counts after 14 days liquid culture of edited and shRNA transduced HSCs. Results of a linear regression model for the effect of edited gene (referent to AAVS1), shRNA (referent to scramble control), and the interaction term of edited gene with shRNA are presented below. Unadjusted p-values from a two-sided test are reported. The horizontal line in each box indicates the median, the tops and bottoms of the boxes indicate the interquartile range, and the top and bottom error bars indicate maxima and minima, respectively. n=4 for AAVS1 gRNA/scramble, n=5 for AAVS1 gRNA/TCL1A shRNA, n=4 for TET2 gRNA/scramble, and n=7 for TET2 gRNA/TCL1A shRNA, which represent biologically independent replicates.