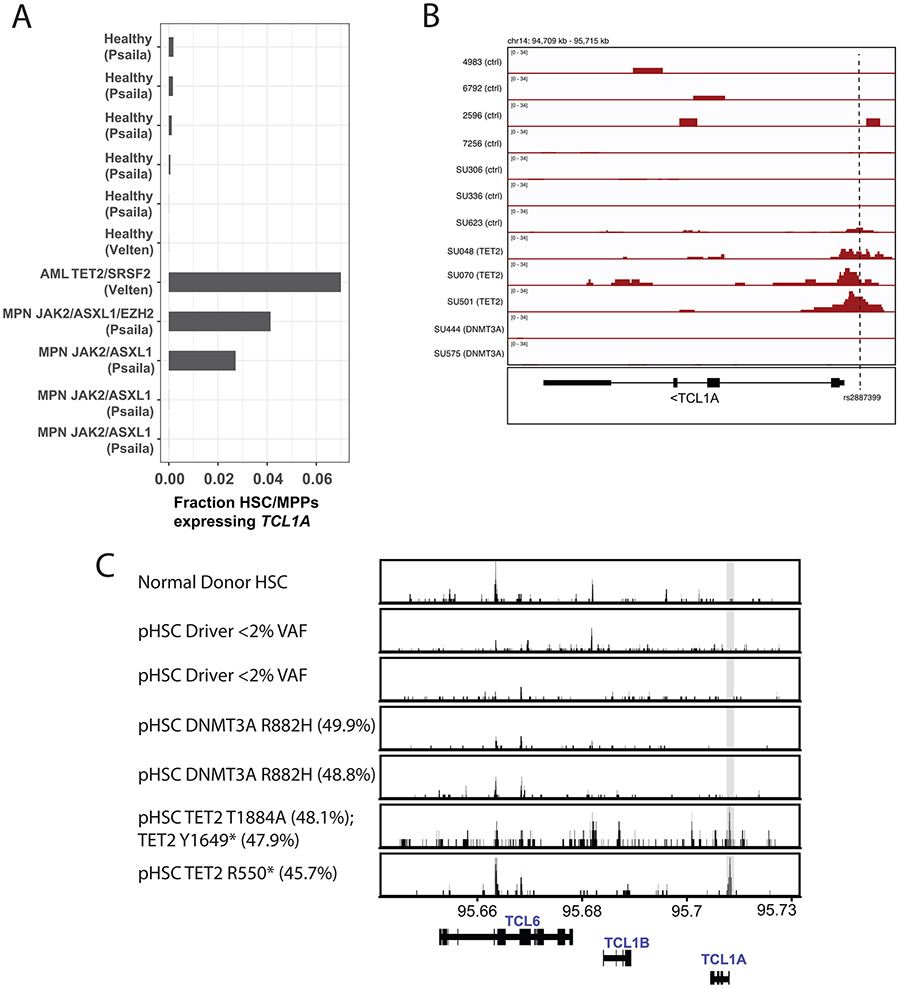
Extended Data Fig 3∣. Chromatin Accessibility and Transcript Expression of TCL1A.
A. Quantification of fraction of HSC/MPPs expressing TCL1A transcripts in patients with TET2 or ASXL1 driven acute myeloid leukemia (AML) or myeloproliferative neoplasm (MPN) compared to healthy donors. Data is from single-cell RNA sequencing generated in Psaila33 et al. and Velten32 et al. B. ATAC-sequencing tracks of the TCL1A locus near rs2887399 in HSCs from healthy donors (row 1-4), pre-leukemic hematopoietic stem cells (pHSCs) from patients with AML but no detected driver mutations (rows 5-7), in pHSCs with TET2 mutations (rows 8-10), and pHSCs with DNMT3A mutations (rows 11-12). Data is from Corces et al36. Vertical dashed line indicates location of the rs2887399 SNP. C. ATAC-sequencing tracks of the TCL6-TCL1A locus in HSCs from healthy donors (row 1), pre-leukemic hematopoietic stem cells (pHSCs) from patients with AML but no detected driver mutations (rows 2-3), pHSCs with DNMT3A mutations (rows 4-5), and in pHSCs with TET2 mutations (rows 6-7). Amino acid change and variant allele fraction (VAF) for the driver mutations are shown. Data is from Corces et al34.