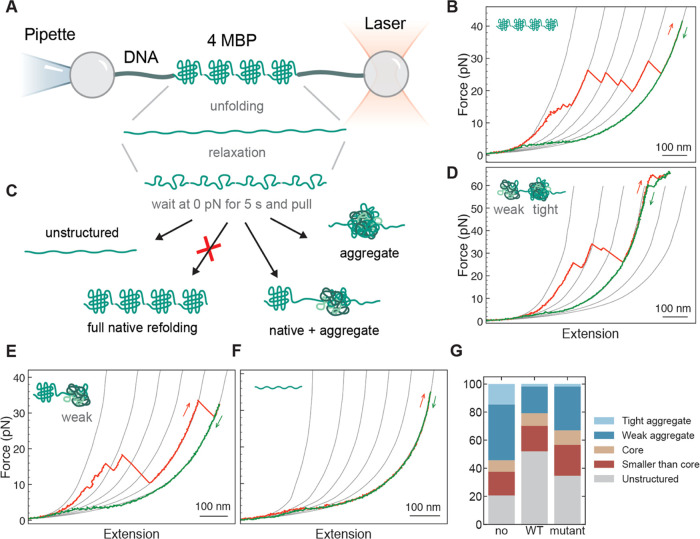
Figure 1.
Mechanical manipulation of 4MBP with and without the presence of chaperones. (A) Four maltose binding proteins arranged in tandem (4MBP) are mechanically manipulated with polystyrene beads by means of DNA molecular handles. One bead is held at the end of a micropipette by suction, while the other is held in an optical trap. By moving the beads relative to each other, the protein can be stretched and relaxed, while the molecular extension and the applied force can be measured as described in refs (21, 64). (B) When stretched for the first time, 4MBP starts losing its structure when external α helices unzip from each monomer and unfold. These structural changes generate a 4MBP lengthening of 100 nm that gives rise to a gradual discontinuity in the stretching trace at ∼10 pN (B). At higher forces (∼25 pN), the remaining core structures unfold sequentially giving rise to a sawtooth-like pattern where each rip corresponds to the unfolding of 250–290 aa, as estimated according to the procedure described in the Data Analysis section, where it is also explained the origin of the reference gray lines. (C) After complete denaturation of the 4MBP molecule, the applied force is relaxed and held at 0 pN for 5 s before the molecule is pulled again. During this relaxation period, amino acids from adjacent domains can interact and end up in different molecular states, as depicted in (C). An analysis of the unfolding jumps observed in the second or subsequent stretching traces allowed us to distinguish 5 molecular states: (i) “tight aggregates”, i.e., compact structures that survive at forces larger than 63 pN (D), (ii) “weak aggregates”, related to jumps that involve more than 290 aa ((D) and (E)), (iii) “core-like structures”, related to jumps that involve between 250 and 290 aa (E), (iv) “small structures”, related to jumps that involve less than 250 aa ((D) and (E)), and (v) “unstructured”, all of the amino acids that do not end up into any of the previous categories (F). (G) Percentage of aa that end up in each of these molecular states in the presence of no chaperone (107 traces; 3 individual molecules), HSPB8 (5 μM) (132 traces; 4 molecules), or HSPB8-K141E (5 μM) (190 traces; 11 molecules).