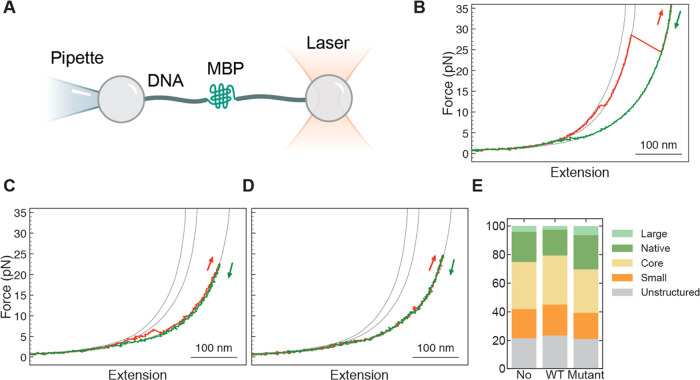
Figure 2.
Mechanical manipulation of sMBP with and without the presence of chaperones. (A) A single sMBP is mechanically manipulated within the same experimental setting employed in 4MBP case (Figure 1A). (B–D) Force vs extension curves of stretching-relaxation cycles performed on an sMBP. The gray lines are the reference curves for the elastic behavior of the handles plus a chain of 0, 91, and 370 unfolded amino acids, from left to right respectively, as explained in the Data Analysis section. While the relaxation curves (green) hardly vary, the stretching (red) curves reveal details about the state of the protein. (B) Unfolding of a native state, characterized by the denaturation of α helices around 10 pN, followed by the unfolding of the core structure around 25 pN. (C) Denaturation of a structure clearly smaller than a typical core. (D) Stretching of an unstructured amino acid chain, without any detectable discrete unfolding events. (E) Each stretching trace can be classified into one of 5 categories (defined in the text), using 179 traces and 8 individual molecules for the dataset without chaperone, 214 traces and 6 molecules for the dataset with wild-type HSPB8, and 155 traces and 5 molecules for the dataset with the mutant chaperone. The observed relative proportion is largely unaffected by the presence of HSPB8, both wild type (5 μM) and mutant (5 μM). Note that this chart is based on a classification of traces, unlike Figure 1G which is based on a classification of jump events.