Figure 1.
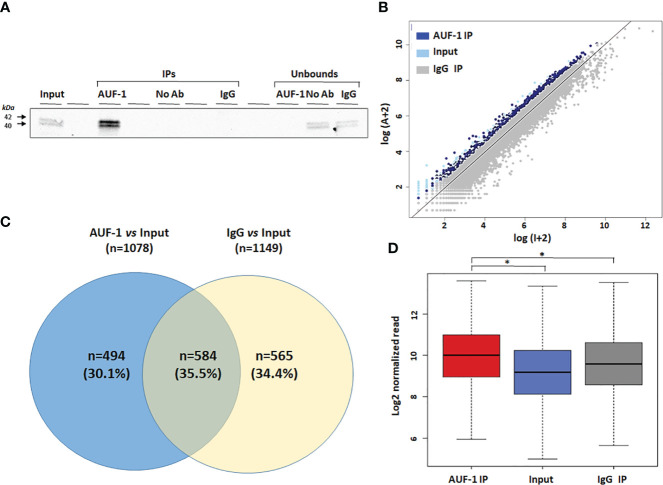
Transcripts associated with AUF-1 in unstimulated BEAS-2B cells identified by RIP-Seq. (A) Representative immunoblot analysis (n=3) showing selective AUF-1 IP compared to unbound controls. The Input samples (Input) were incubated with both AUF-1 and IgG antibodies (AUF-1 IP and IgG IP, respectively) and then immunoprecipitated with magnetic beads. An additional antibody-free control sample (no Ab) was performed. (B) Scatter plot of RIP-Seq data. Read counts for AUF-1 IP, IgG and Input controls were normalized and log-transformed. Dark blue and light blue dots represent enriched AUF-1 IP and IgG targets (EF≥1.5 and FDR ≤ 0.05), respectively. Gray dots represent background (Input). Axes represent log2 read count in Input (X) and AUF-1 IP (Y). (C) Venn diagram showing exclusive and overlapping targets between AUF-1 IP- and IgG-IP-enriched transcripts (each total number in parenthesis) vs Input (EF ≥ 1.5 and FDR ≤ 0.05). (D) Boxplot showing the enrichment of the 494 AUF-1 transcript targets in Input, AUF-1 IP and IgG IP samples. Y axis represents the log2 of the normalized read count. *p ≤ 0.05 (Student’s t-test).