Figure 2.
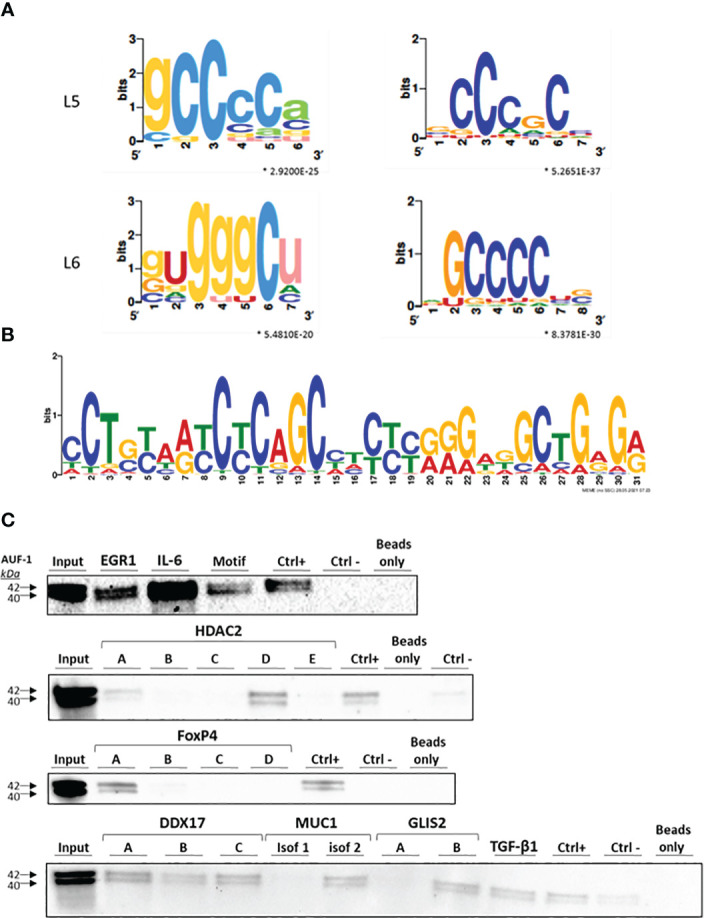
Identification of predicted binding motifs in AUF-1-associated transcripts. (A) k-mer length 5 (L5) and 6 (L6) graphic generated using the SMARTIV tool representing the probability matrix of the AUF-1 motif, showing the relative frequency of each nucleotide for each position within the motif sequence. The motif originated from the 3’UTR sequences of the exclusive n= 494 AUF-1 transcripts ( Figure 1C ) obtained from the RIP-Seq study. Upper and lower case letters show the secondary structure prediction (A, G, C, U for unpaired nucleotides and a,g,c,u for paired nucleotides). *p-value ≤ 0.05 according to minimum hypergeometric statistical approach. (B) Graphic of the 30-mer consensus AUF-1-binding motif generated using the MEME tool, identified within the top 1,000 peaks originating from the 3’UTR sequences of RIP-Seq – derived AUF-1 targets (full list of consensus motifs in Figure S2 ). This motif was utilized to generate the biotinylated probe for validation with biotin pull-down reported in panel C (listed in Table 2 ). (C) Representative immunoblots (n=2) of AUF-1 detection by RNA biotin pulldown in BEAS-2B cytoplasmic lysates using biotinylated 3’UTR probes for the indicated AUF-1 targets and the in vitro biotinylated AUF-1 GC-rich motif, synthesized from consensus sequence shown in panel B, selecting highest-frequency nucleotides. Capital letters (A to E) represent biotinylated fragments of adjacent sequences used for long 3’UTRs; “CTRL+”, positive control (Cyclin D1 3’UTR); “CTRL-”, negative control (PD-L1 coding sequence).
