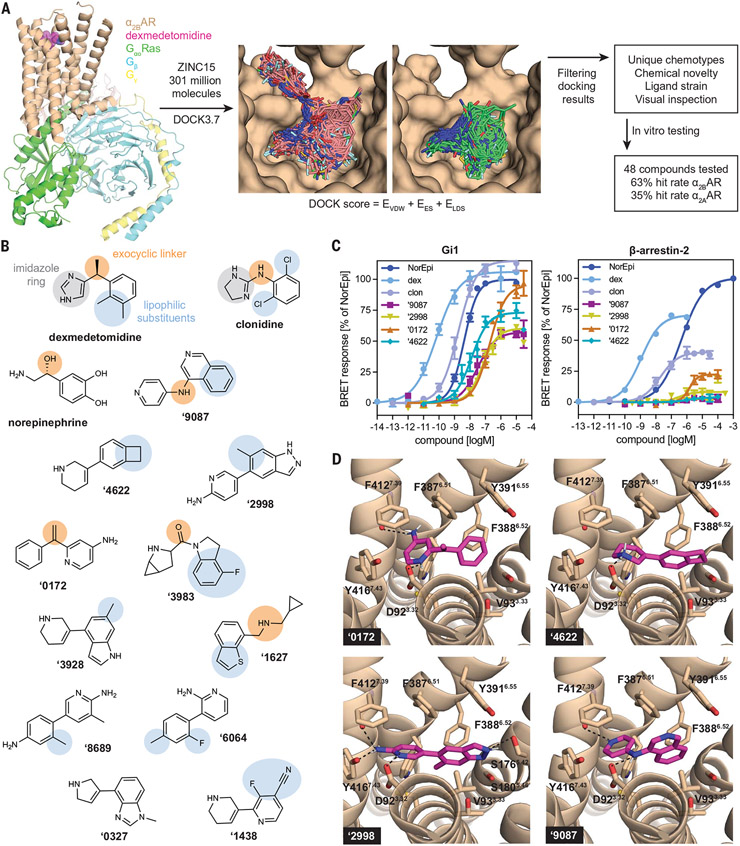
Fig. 1. Newly discovered a2AAR agonists from ultralarge library docking.
(A) 301 million molecules were docked against the active state of a2BAR (PDB 6K41). Lead-like molecules (pink carbons) often spilled out of the orthosteric site, whereas fragment molecules (green carbons) are well complemented by that site. Hit rates were determined with a Ki cutoff of 10 μM. EVDW, van der Waals; EES, electrostatic; ELDS, ligand desolvation. (B) The αAR pharmacophore model (9) overlaid on known α2AAR agonists dexmedetomidine, clonidine, and norepinephrine and new agonists from docking (colors represent the different moieties fulfilling the same role). (C) Gi activation and β-arrestin-2 recruitment for norepinephrine (NorEpi), dexmedetomidine (dex), clonidine (clon), and several of the newly discovered docking agonists. (D) Docked poses of these new agonists with hydrogen bonds to key recognition residues of α2BAR shown as black dashed lines. For (C), data are means ± SEMs of normalized results (n = 4 to 17 measurements for Gi and n = 3 to 8 measurements for β-arrestin-2). Single-letter abbreviations for the amino acid residues are as follows: A, Ala; C, Cys; D, Asp; E, Glu; F, Phe; G, Gly; H, His; I, Ile; K, Lys; L, Leu; M, Met; N, Asn; P, Pro; Q, Gln; R, Arg; S, Ser; T, Thr; V, Val; W, Trp; and Y, Tyr.