Abstract
Background & Aims:
We recently reported use of tissue-based transcriptomic biomarkers (miRNA or mRNA) for identification of lymph node metastasis (LNM) in patients with invasive submucosal colorectal cancers (T1 CRC). In this study, we translated our tissue-based biomarkers into a blood-based liquid biopsy assay for noninvasive detection of LNM in patients with high-risk T1 CRC.
Methods:
We analyzed 330 specimens from patients with high-risk T1 CRC, which included 188 serum samples from two clinical cohorts (training cohort: n=46, validation cohort: n=142) and matched FFPE samples (n=142). We performed RT-qPCR followed by logistic regression analysis to develop an integrated transcriptomic panel and establish a risk-stratification model, combined with clinical risk factors.
Results:
We used comprehensive expression profiling of a training cohort of LNM-positive and -negative serum specimens to identify an optimized transcriptomic panel of four miRNAs (miR-181b, miR-193b, miR-195, miR-411) and five mRNAs (AMT, FOXA1, PIGR, MMP1, MMP9), which robustly identified patients with LNM (area under the curve [AUC]=0.86, 95% CI=0.72–0.94). We validated panel performance in an independent validation cohort (AUC=0.82, 95% CI=0.74–0.88). Our risk-stratification model was more accurate than the panel and an independent predictor for identification of LNM (AUC=0.90, Univariate: odds ratio [OR]=37.17, 95% CI=4.48–308.35, P<.001; Multivariate: OR=17.28, 95% CI=1.82–164.07, P=.013). The model limited potential overtreatment to only 18% of all patients, which is dramatically superior to currently used pathological features (92%).
Conclusions:
A novel risk-stratification model for noninvasive identification of T1 CRC has the potential to avoid unnecessary surgeries for patients classified as high-risk by conventional risk-classification criteria.
Keywords: transcriptomic panel, risk-stratification model, detection biomarker, noninvasive assay
LAY SUMMARY
This study reports a novel biomarker signature that can distinguish high-risk T1 CRCs patients wi th lymph node metastasis from those who do not.
INTRODUCTION
In recent years, the diagnosis of invasive submucosal colorectal cancers (T1 CRCs) has increased by up to 15–30% due to the implementation of mass CRC screening and frequent patient examinations.1, 2 However, recent advances in endoscopic devices have enabled curative treatment via endoscopic submucosal dissection (ESD) or endoscopic mucosal resection (EMR), for patients with T1 CRC who would have otherwise been treated by radical surgeries.3 This has prompted the National Comprehensive Cancer Network to recommend ESD as a preferred treatment modality for patients with suspected T1 CRC. Successful treatment of patients with T1 CRCs starts with accurate diagnosis during endoscopy. However, two prospective studies recently highlighted that 30–40% of these patients are misdiagnosed, and the pre-surgical discrimination of T1 CRC remains clinically challenging.4, 5 Although some patients can be successfully treated with ESD or EMR, approximately 70–80% of patients with T1 CRC require radical surgeries to achieve a complete cure, due to the potential risk for lymphnode metastasis (LNM) after pathological analysis, which is estim ated to occur in as many as 5–15% of patients with high-risk T1 CRC.6–8
With the implementation of endoscopic treatment for suspected T1 CRCs, it has become necessary to identify the risk of LNM, in order to select patients who truly have high-risk disease and require radical surgery, while sparing others from overtreatment. The currently used pathological criteria to identify LNM in patients with T1 CRC include depth of submucosal invasion (>1000 μm), presence of lymphatic or vascular invasion, high-grade tumor budding, and poorly differentiated histology.9–13 If these factors are absent, endoscopic treatment is considered sufficient to cure patients with T1 CRC who have low-risk for LNM.14, 15 Unfortunately however, in clinical settings, if even one of these pathological risk features is present, the patient is deemed as “high risk for LNM” and is recommended to undergo additional surgery.3, 11, 16, 17 Such a dichotomized clinical management approach for patients with T1 CRCs has serious drawbacks, as it often leads to overtreatment, even though the actually have LNM.10, 14, 16, 19–25 This highlights an important clinical challenge: we need more prudent risk assessment for limiting unnecessary radical surgery in 85–95% of patients with T1 CRC. In addition, these data suggest the inadequacy of currently used pathological risk factors and emphasize the need to develop robust molecular biomarkers that can identify the presence of LNM pre-operatively, which would better inform clinical decision-making in patients with T1 CRC, minimize the number of surgeries performed, and reduce the overall burden of costs associated with such invasive surgeries.
Accumulating evidence indicates that the expression pattern of microRNAs (miRNAs) reflects the physiological and pathological status of cancer patients. In fact, several studies have identified the differential expression of specific miRNAs to be directly involved in CRC pathogenesis, as well as emphasized their potential as circulating biomarkers for CRC.26–30 Although considerable advances have been made in exploiting miRNAs as noninvasive diagnostic biomarkers,31–33 using circulating miRNAs to identify high-risk T1 CRCs clinically has thus far not been attempted. We previously described a panel of tissue-based miRNAs and gene expression biomarkers that allowed robust detection of LNM in patients with T1 CRC.34, 35 However, an ideal clinical application of these biomarkers would be to use them to diagnose patients with high-risk T1 CRC prior to surgery, before such tissue specimens are readily available. Therefore, translating these biomarkers into a “liquid biopsy” assay is attractive, as this would allow a noninvasive, facile, and inexpensive diagnostic assay for LNM in patients with high-risk T1 CRC. To address this gap in knowledge, we evaluated the feasibility of translating our previously reported transcriptomic biomarkers (miRNAs and mRNAs), into a blood-based, noninvasive assay by systematically analyzing blood specimens from multiple cohorts of patients with T1 CRC. As a result, we successfully established a novel, blood-based, transcriptomic signature that robustly identified the presence of LNM in patients with T1 CRC, with an area under the curve (AUC) value of 0.90. This assay allowed reclassification of 75% of high-risk T1 CRCs into the low-risk group, which would obviate the need for unnecessary surgeries in this significant majority of patients who would have otherwise been subjected to radical surgeries based upon conventional pathological risk-assessment criteria.
MATERIALS AND METHODS
Patient cohorts
We analyzed a total of 330 patient samples, which included 188 serum specimens from patients with high-risk T1 CRCs comprised of 2 independent clinical cohorts: a training cohort (n=46; 5 LNM-positive (LNP) and 41 LNM-negative (LNN) patients) from the National Cancer Center Hospital, Japan; and a validation cohort (n=142; 12 LNP and 130 LNN patients) from the National Cancer Center Hospital East, Japan (Fig. 1A). Matched formalin-fixed paraffin-embedded (FFPE; n=142) specimens, which were obtained following endoscopic or surgical tumor resection, were also obtained from patients within the validation cohort. All patients were diagnosed as “high-risk” pathologically. The pathological criteria included depth of submucosal invasion (>1000 μm), presence of lymphatic or vascular invasion, high-grade tumor budding, and poorly differentiated histology. All patients underwent radical surgeries between January 2017 and December 2017 in the training cohort, and between January 2011 and December 2017 in the validation cohort. Exclusion criteria were: synchronous advanced CRCs, presence of distant metastases, hereditary or inflammation-associated CRC, non-adenocarcinoma, or non-availability of serum specimens.
Figure 1.
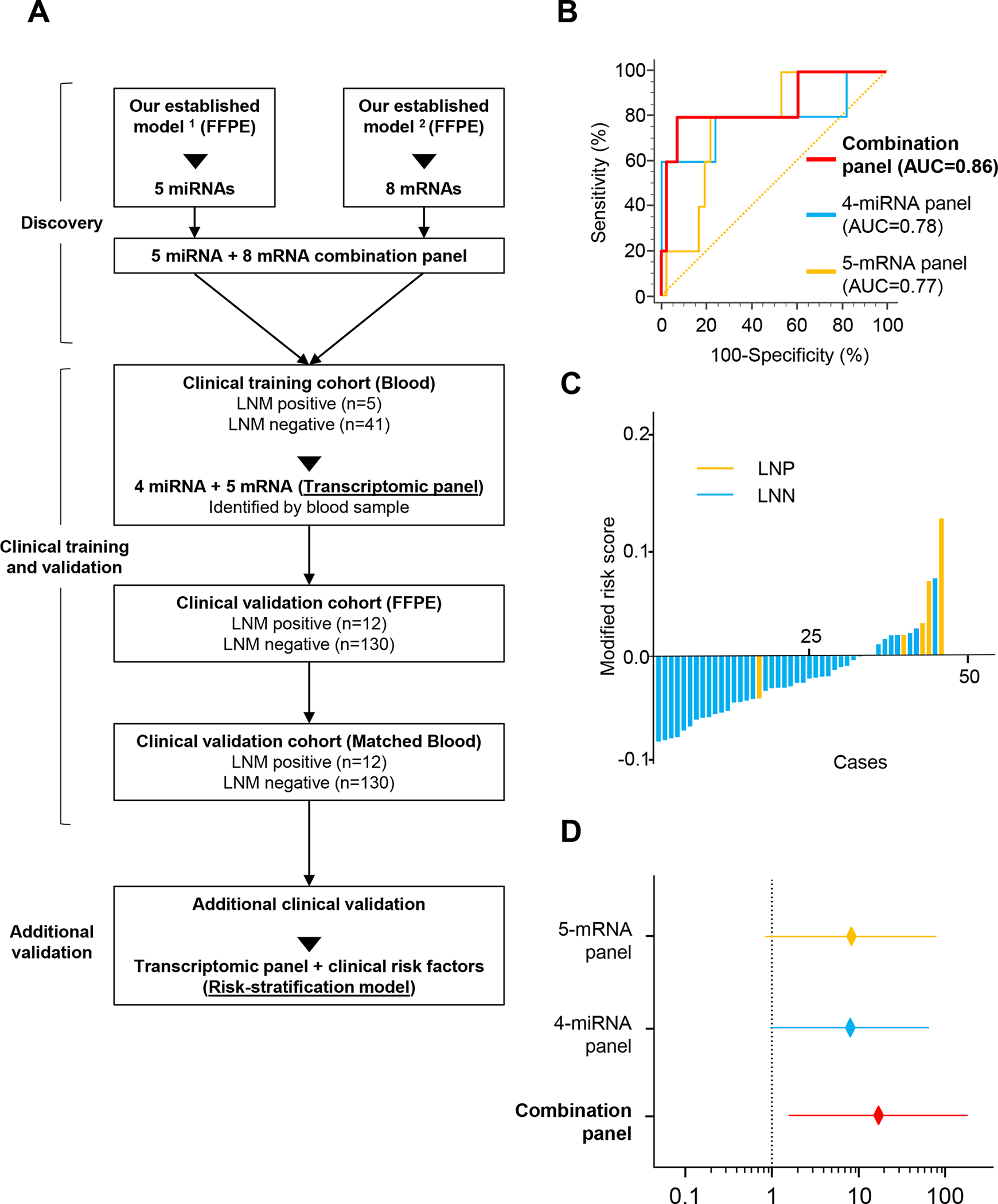
Training phase of a transcriptomic panel for the identification of LNM in patients with T1 CRC. A) Overview of the study. B) A ROC curve for a 4-miRNA and 5-mRNA panel in serum from training cohort patients (LNP = 5, LNN = 41; AUC: 4-miRNA panel = 0.78, 5-mRNA panel = 0.77, combination panel = 0.86). C) Risk score distribution plot in training cohort patients. Modified risk scores were obtained from individual risk scores by using Youden’s index values from the risk model. D) Forest plots with ORs for each panel risk score status in univariate logistic regression analysis in training cohort patients (ORs: 4-miRNA panel = 8.62, 5-mRNA panel = 8.44, combination panel = 14.22).
All patients underwent standard endoscopic and surgical procedures (resection of affected segment of colon or rectum and regional lymphadenectomy), and all specimens were evaluated by pathologists at each participating institution, according to the 7th edition of the American Joint Committee on Cancer TNM grading system. The study was conducted in accordance with the Declaration of Helsinki. Written informed consent was obtained from all patients, and the study was approved by the institutional review boards of all participating institutions.
RNA extraction from serum and FFPE specimens
Total RNA extraction from all serum specimens was performed using the Qiagen miRNeasy Kit (Qiagen, Hilden, Germany). Briefly, 200 μL of serum was thawed on ice and centrifuged at 3000 ×g for 5 min to remove cell debris. Next, 200 μL of the supernatant was lysed in 5 times the volume of Qiazol solution. To normalize any inadvertent sample-to-sample variations during the characteristics were compared between LNP and LNN patients, using the Chi-Square test or Mann-Whitney U test for categorical data. Binary logistic regression was used to train a classifier based on the expression of four miRNAs and five mRNAs. Of note, once the model was trained in the training cohort, the same statistical model variables (weights and cutoff thresholds) were applied in the validation cohort. The LNM risk score for all patients was calculated based on the individual biomarker coefficients derived from the training cohort as follows: Logit (P) = (−0.318*MIR181b) + (−0.762*MIR193b) + (−1.019*MIR195) + (−0.627*MIR411) + (−0.135*AMT) + (0.010*FOXA1) + (0.241*MMP1) + (−0.776*MMP9) + (0.231*PIGR) − 8.363. The cutoff threshold for the LNM risk score was chosen as 0.08, which was determined by Youden’s index. For all cohorts, receiver operator characteristic curves and AUC values were used to evaluate the performance of the panel for LNM detection in patients with T1 CRC. A P value <0.05 was considered statistically significant. Statistical analyses were performed using JMP Genomics V 9.0 statistical software (SAS Institute Japan, Tokyo, Japan), Medcalc statistical software V.16.2.0 (Medcalc Software bvba, Ostend, Belgium), GraphPad Prism V7.0 (GraphPad Software, San Diego, CA, USA), and R (3.5.0, R Development Core Team, https://cran.r-project.org/).
RCC1) were not detectable in serum specimens, which led us to establish a panel of four miRNAs (miR-181b, miR-193b-3p, miR-195–5p, and miR-411–5p) and five mRNAs (AMT, FOXA1, MMP1, MMP9, and PIGR). Next, we systematically interrogated the diagnostic accuracy of our transcriptomic panel for its ability to detect LNM in patients with T1 CRC. Using logistic regression analysis, we trained a risk-assessment model in the training cohort of patients that allowed robust identification of LNM in patients with T1 CRC using the four miRNAs (AUC = 0.78, 95% CI = 0.64–0.89) or the five mRNAs (AUC= 0.77, 95% CI = 0.62–0.88) (Fig. 1B, C and Supplementary Table 2). Identification of LNM was notably superior when we used all four miRNAs and five mRNAs to establish a combined transcriptomic panel (AUC = 0.86, 95% CI = 0.72–0.94).
We performed univariate analysis to confirm that each of the biomarker panels was quite robust individually (miRNA panel: odds ratio [OR] = 8.62, P = .06; mRNA panel: OR = 8.44, P = .05; Fig. 1D, Supplementary Table 3). However, the combined panel was significantly superior in diagnosing the presence of LNM in patients with T1 CRC (OR = 14.22; P = .02). We developed this risk-assessment scoring model based on the coefficients derived from individual the diagnostic accuracy of our transcriptomic panel in these FFPE surgical specimens, and were enthused to observe that its diagnostic performance was comparable to that observed in serum specimens in the training cohort (AUC = 0.83, 95% CI = 0.75–0.89; Fig. 2A). When we evaluated the performance of the signature in matched blood serum specimens, the diagnostic accuracy was in line with the findings from tissue specimens (AUC = 0.82, 95% CI = 0.74–0.88, Fig. 2B, C). This highlights the clinical significance of our transcriptomic panel in identifying presence of LNM in patients with T1 CRC.
Figure 2.
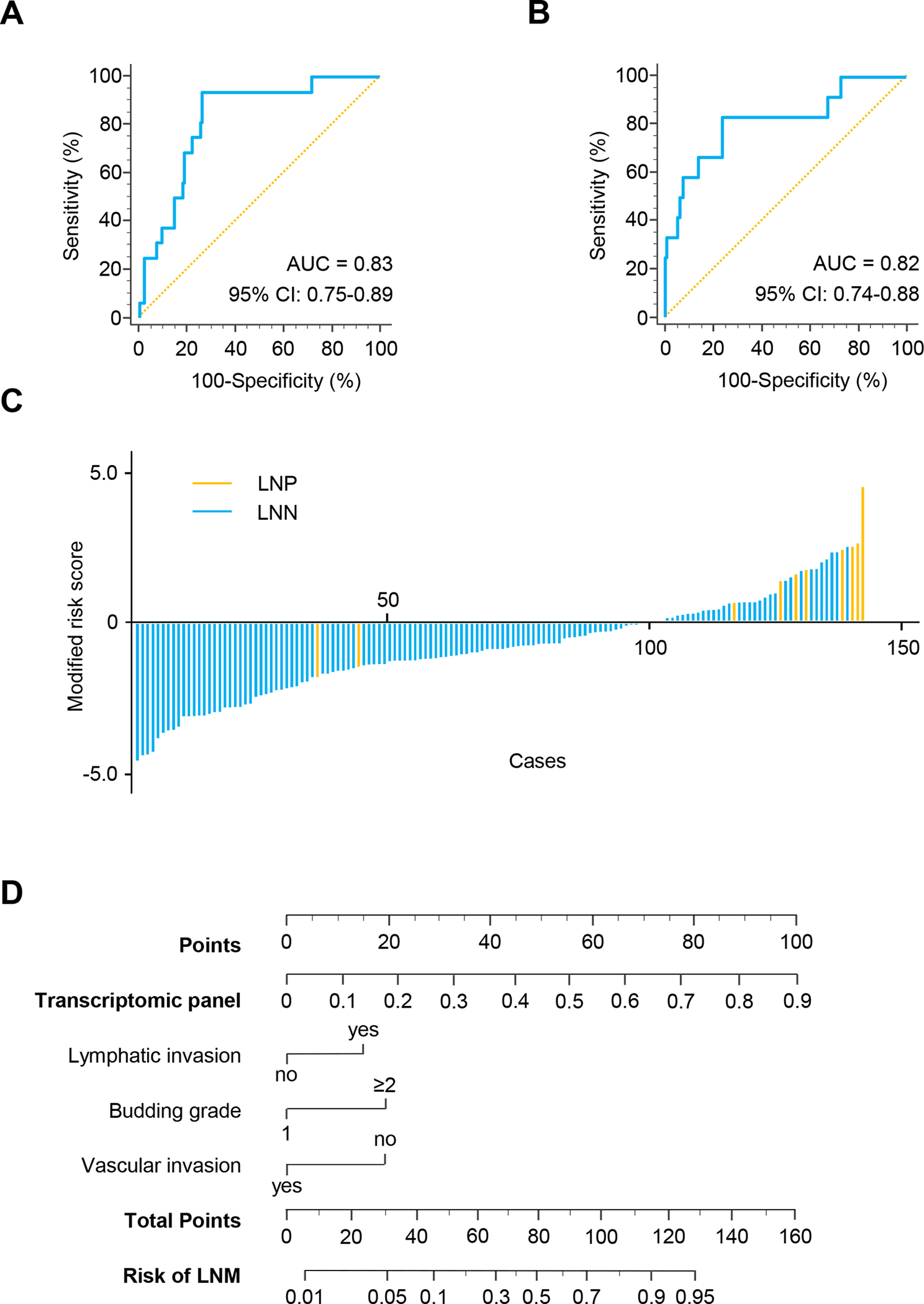
Validation phase of the transcriptomic panel for the identification of LNM in patients with T1 CRC. A) A ROC curve for the transcriptomic panel in tissue specimens from validation cohort patients (LNP = 12, LNN = 130, AUC = 0.83). B) A ROC curve for the transcriptomic panel in matched serum samples in validation cohort patients (LNP = 12, LNN = 130, AUC = 0.82). C) Risk score distribution plot in serum specimens from validation cohort patients. D) A nomogram illustrating the probability of LNM risk. For clinical purposes, the scores of each covariate are added, and the total score is depicted on the total score point axis.
For an easier translation of the biomarker panel into the clinic, we evaluated its performance along with other pathological risk features (i.e. lymphatic or vascular invasion, high-grade tumor budding), and established a nomogram for predicting the diagnostic probability for the presence of LNM from validation cohort. Through ranking the effect estimators, point scores were assigned to each risk factor. The total points accumulated from all the risk factors corresponded to the predicted probability of LNM for individual patients. We incorporated all pathological and molecular risk features and determined that although other pathological risk-assessment features added some weight to the model, our panel had the highest weight in this model and was an independent and the most significant predictor for the presence of LNM in patients with T1 CRC (Fig. 2D).
A risk-stratification model that combines transcriptomic biomarkers and current risk-assessment features significantly improves diagnosis of LNM in patients with T1 CRC Considering the current landscape of widely used clinical risk factors for identifying patients with T1 CRC, we asked whether a risk-stratification model that includes some of the currently used pathological risk features (i.e. lymphatic and vascular invasion, tumor budding grade, and depth of tumor invasion) along with our transcriptomic biomarkers might further improve diagnostic accuracy in detecting LNM in patients with T1 CRC. As 12 patients were lack of clinical information, totally 130 patients were included in risk-stratification model. When we performed such an analysis in the patients within the serum specimens of validation cohort, this led to a significant improvement in its diagnostic sensitivity and specificity for the identification of LNM (AUC = 0.90, 95% CI = 0.83–0.95, Fig. 3A and Table 2).
Figure 3.
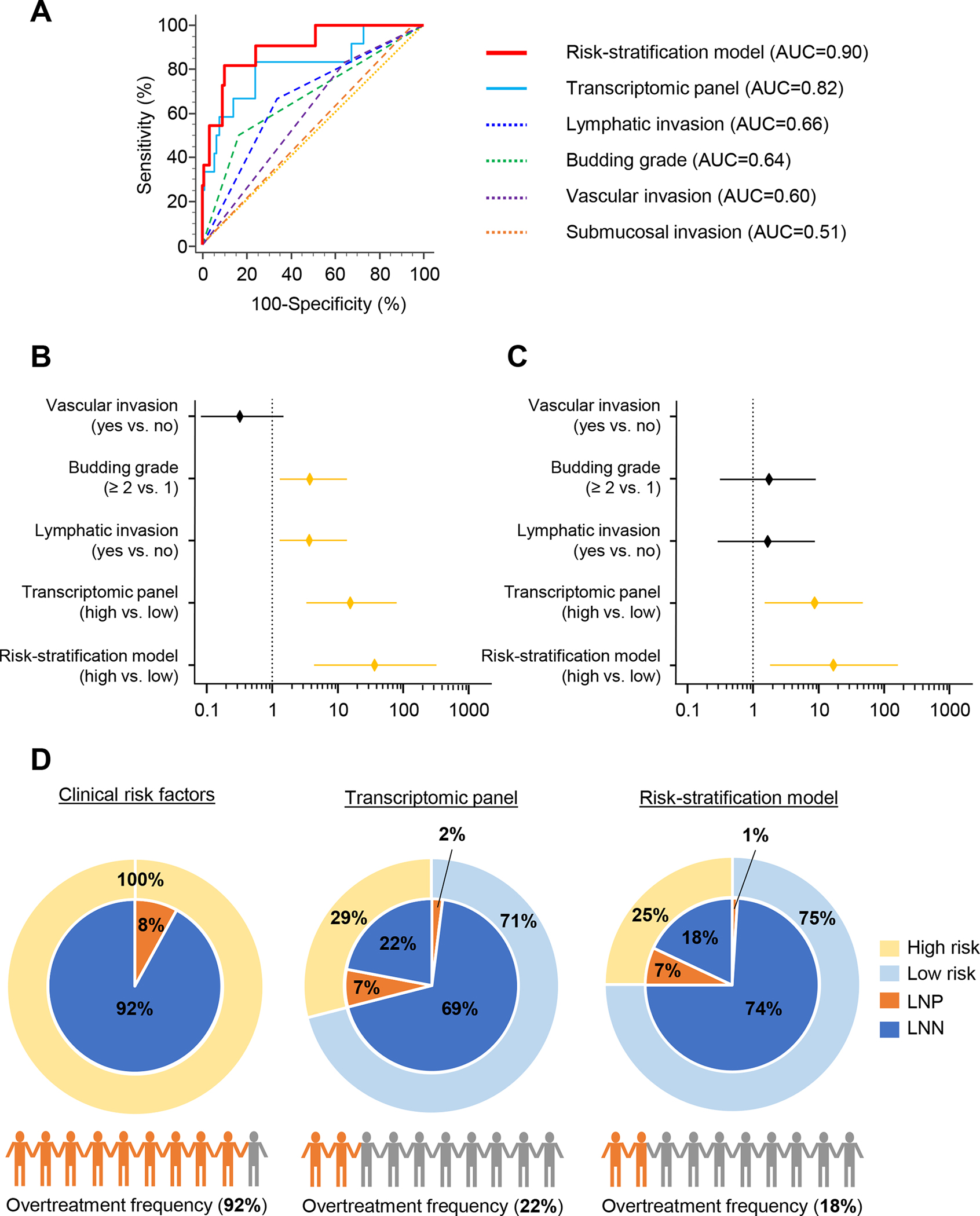
Clinical validation of the risk-stratification model in patients with T1 CRC. A) The risk-stratification model, which combines the transcriptomic panel and pathological risk factors, outperformed detection accuracy of the transcriptomic panel or risk factors alone in serum specimens from validation cohort patients (AUC = 0.90). B–C) Forest plot with ORs of clinicopathological variables, transcriptomic panel, and risk-stratification model in univariate (B) and multivariate (C) logistic regression analysis in validation cohort patients. D) Currently used pathological factors led to the overtreatment of 92% patients with T1 CRC (left panel). The patients in validation cohort using our transcriptomic classifier divided into high (Yellow) and low (Light blue) risk by Youden’s index. Pie chart shows LNM status of LNP (Orange) and LNN (Dark blue). The transcriptomic panel would have led to the overtreatment of only 22% patients with T1 CRC (middle panel), and the risk-stratification model would have led to the overtreatment of only 18% patients with T1 CRC (right panel).
Table 2.
Model Performance in Estimating the Risk of Lymph Node Metastasis
| Value (95% CI) | ||||
|---|---|---|---|---|
|
|
||||
| Variable | Training cohort (Blood) | Validation cohort (FFPE) | Validation cohort (Blood) | Risk-stratification model (Blood) |
|
| ||||
| Cutoff value | 0.08 | 0.05 | 0.08 | 0.08 |
| Sensitivity, % | 80.0 (28.4–99.5) | 91.7 (61.5–99.8) | 83.3 (51.6–97.9) | 90.0 (55.5–99.7) |
| Specificity, % | 92.7 (80.1–98.5) | 73.9 (65.4–81.2) | 76.2 (67.9–83.2) | 81.4 (73.1–87.9) |
| AUC, % | 85.5 (71.8–94.0) | 82.6 (75.4–88.5) | 81.5 (74.1–87.5) | 90.0 (83.4–94.6) |
| PPV, % | 57.1 (29.2–81.2) | 24.4 (18.8–31.2) | 24.4 (17.8–32.4) | 29.0 (21.0–38.6) |
| NPV, % | 97.4 (86.8–99.5) | 99.0 (93.6–99.8) | 98.0 (93.3–99.4) | 99.0 (93.7–99.8) |
NPV, negative predictive value; PPV, positive predictive value.
We next determined specific diagnostic correlates for our combined biomarker panel in blood samples from the validation cohort; its sensitivity, specificity, PPV, and negative predictive value (NPV) were 83.3%, 76.2%, 24.4%, and 98.0%, respectively (Table 2). When we performed a similar analysis of the newly established risk-stratification model that also included pathological risk features, its performance was significantly superior; its sensitivity, specificity, PPV, and NPV were 90.0%, 81.4%, 29.0%, and 99.0%, respectively. This highlights the superiority of the risk-stratification model for identifying LNM in patients with T1 CRC.
We next categorized all patients into high- and low-risk groups using cutoff thresholds derived from Youden’s index for the nine miRNA and mRNA biomarkers. Accordingly, we performed univariate logistic regression analysis, which revealed that our transcriptomic panel emerged as an independent predictor for LNM in patients with T1 CRC in both clinical cohorts, compared to any singular clinical risk factor (training cohort: OR = 14.22, 95% CI = 1.41–143.68, P = .025; validation cohort: OR = 15.97, 95% CI = 3.32–76.82, P < .001; Table 3). Further, univariate and multivariate logistic regression analysis revealed that our novel risk-stratification model was a superior than the panel and independent predictor of LNM (Univariate: OR = 37.17, 95% CI = 4.48–308.35, P < .001; Multivariate: OR = 17.28, 95% CI = 1.82–164.07, P = .013) in the validation cohort of patients (Fig. 3B, C and Table 3). Collectively, these data highlight the potential clinical significance of our risk-stratification model for diagnosis and risk assessment in the identification of LNM.
Table 3.
Univariate and Multivariate Logistic Regression Analysis for Lymph Node Metastasis
| Univariate analysis | Multivariate analysis | |||||
|---|---|---|---|---|---|---|
|
|
|
|||||
| Factors | OR | 95% CI | P a | OR | 95% CI | P a |
|
| ||||||
| Training cohort (N = 46) | ||||||
| Age | ||||||
| (≥67 vs <67) | 0.22 | 0.02–2.10 | .19 | |||
| Sex | ||||||
| (male vs female) | 0.58 | 0.09–3.82 | .57 | |||
| Tumor location | ||||||
| (right vs left) | 0.39 | 0.04–3.82 | .42 | |||
| Tumor size | ||||||
| (≥20 mm vs <20 mm) | 1.58 | 0.24–10.44 | .64 | |||
| Submucosal invasion | ||||||
| (≥1000 μm vs <1000 μm) | <0.01 | .99 | ||||
| Budding grade | ||||||
| (≥2 vs 1) | 3.57 | 0.42–30.10 | .24 | |||
| Lymph invasion | ||||||
| (positive vs negative) | 1.82 | 0.27–12.38 | .54 | |||
| Vascular invasion | ||||||
| (positive vs negative) | 4.65 | 0.68–31.91 | .12 | |||
| Transcriptomic panel | ||||||
| (high risk vs low risk) | 14.22 | 1.41–143.68 | .025 | |||
| Validation cohort (N = 142) | ||||||
| Age | ||||||
| (≥67 vs <67) | 0.97 | 0.30–3.16 | .96 | |||
| Sex | ||||||
| (male vs female) | 1.33 | 0.38–4.66 | .65 | |||
| MSI status | ||||||
| (MSI-H vs MSI-L, MSS) | 1.22 | 0.14–10.56 | .86 | |||
| Tumor location | ||||||
| (right vs left) | 0.81 | 0.21–3.15 | .76 | |||
| Tumor size | ||||||
| (≥20 mm vs <20 mm) | 7.82 | 0.98–62.35 | .05 | |||
| Submucosal invasion | ||||||
| (≥1000 μm vs <1000 μm) | <0.01 | .99 | ||||
| Budding grade | ||||||
| (≥2 vs 1) | 3.89 | 1.08–13.95 | .037 | 1.70 | 0.32–9.06 | .53 |
| Lymph invasion | ||||||
| (positive vs negative) | 3.78 | 1.08–13.23 | .038 | 1.60 | 0.29–8.69 | .59 |
| Vascular invasion | ||||||
| (positive vs negative) | 0.35 | 0.07–1.68 | .19 | |||
| Transcriptomic panel | ||||||
| (high risk vs low risk) | 15.97 | 3.32–76.82 | <.001 | 8.13 | 1.43–46.29 | .018 |
| Risk-stratification model | ||||||
| (high risk vs low risk) | 37.17 | 4.48–308.35 | <.001 | 17.28 | 1.82–164.07 | .013 |
MSI, microsatellite instability; MSI-H, high-frequency microsatellite instability; MSI-L, low-frequency microsatellite instability; MSS, microsatellite stable
Bold P values are statistically significant (P < .05).
Our noninvasive risk-assessment model is significantly superior to currently used pathological risk factors for identifying patients with high-risk T1 CRC and reducing the burden of unnecessary surgical treatments
The ultimate goal of our study was to determine the clinical usefulness of our transcriptomic panel in noninvasively identifying patients who truly have LNM and sparing the rest from unnecessary surgeries. In this study, we only enrolled patients who were deemed high-risk based upon the currently used pathological risk factors. However, only 8% of “high-risk” patients (12 of 142) were actually high risk, indicating that 92% of patients (130 of 142) were erroneously categorized as high risk and underwent unnecessary radical surgeries (Fig. 3D, left panel). In contrast, when we analyzed the same patients using our transcriptomic classifier and divided into high and low risk by Youden’s index, it stratified 29% of patients into the high-risk category (41 of 142). Among these, 10 patients had LNM (7%), indicating that only 22% of the entire cohort (31 of 142) received overtreatment, which is notably superior to potential overtreatment compared with the currently used pathological features (92% vs 22%; Fig. 3D, middle panel). Our newly established risk model was even more accurate than the panel, as it stratified only 25% of patients into the high-risk group (32 of the 130), and the remaining 75% (98 of the 130) of patients were deemed as low risk. Of the 32 patients who were classified as high risk, 9 patients had LNM (7%), indicating that only 18% (23 of 130) of all patients with T1 CRC were potentially overtreated, which is dramatically superior compared with currently used pathological features (92% vs 18%; Fig. 3D, right panel). This highlights the potential for using our liquid biopsy-based risk-assessment model in patients with high-risk T1 CRC.
DISCUSSION
The presence of LNM is an important risk factor for additional surgery following curative endoscopic treatment in patients with T1 CRC. Our present study overcomes the inadequacy of clinicopathologic risk features that are currently used in the clinic to identify LNM in “high-risk” subsets of patients with T1 CRC. Our data demonstrate that a blood-based, transcriptomic assay can be used to accurately estimate risk in pre-operative settings, has a tremendous clinical potential for more robust risk-stratification for the identification of LNM, and can lead to a dramatic reduction in the number of unnecessary surgeries that are currently being performed in these patients. Identifying true high-risk patients and saving others from such unnecessary treatment will reduce patient complications, physician burdens, and associated healthcare costs.37, 38, 39
In this study, our newly established noninvasive risk model exhibited a significantly superior diagnostic accuracy for LNM (AUC= 0.90) vs. the currently used clinical risk models (AUC= 0.73 [training] and 0.76 [validation]) (Supplementary Fig. 1). Although all patients enrolled in our study were deemed to be high-risk for LNM and received radical surgery, post-surgical pathological analyses identified that only 9% (17 of 188 (46 in training cohort and 142 in validation cohort)) of patients were LNP and 91% of patients underwent unnecessary surgeries. Our newly established diagnostic signature revealed that only 18% were overtreated, which is dramatically better for identification of LNM.
Several reports have indicated the potential of ESD for diagnosing LNM in patients with T1 CRC;40, 41 however, others suggest its diagnostic accuracy for LNM is still inadequate.42 Because current clinical guidelines consider the presence of LNM an important risk factor for classifying a patient with T1 CRC as high risk, this highlights the need to develop robust biomarkers for LNM prior to treatment, which would be clinically transformative in selecting patients who truly require such invasive and radical surgical treatments. Our ability to successfully validate our signature in pre-treatment serum samples underscores its clinical significance for improved treatment strategies in patients with T1 CRC, especially the ones who truly have LNM. Our previous studies similarly highlighted the clinical use of pre-treatment serum samples for diagnostic purposes in patients with CRC; however, none of the previous studies used these samples directly for diagnosing LNM status, which could have a profound impact in the selection of treatment strategies.31–33, 43 Pre-operative application of our transcriptomic biomarkers as a robust, facile, and inexpensive assay will lead to minimized risks from surgical procedures, including perforation or bleeding, and a reduction in overall healthcare burden from such expensive surgical procedures.
Our study has some potential limitations, because our retrospective study design might result in a potential selection bias. First, due to the limited sample size (especially small number of positive cases) in the present study, we evaluated our signature in a moderately sized clinical cohort. Thus, a prospective clinical trial with larger patient cohorts is required to further confirm the diagnostic accuracy of our risk-stratification model. Second, our study used training and validation cohorts of patients from Japan, who showed similar clinicopathologic characteristics; such characteristics could potentially vary if we were to analyze patient populations from other countries. Therefore, it will be important to validate the selected biomarkers and our risk-stratification model in patient cohorts from other countries to further reinforce the generalizability of our findings. Finally, we established the risk-stratification model which included miRNAs, mRNAs, and clinical factors. However, previous reports showed that the patients with the consensus molecular subtypes (CMS) and DNA mutations were related to the risk for LNM.44,45 Because fewer factors have the potential for an easier clinical application, future studies may need to explore other factors like CMS or DNA mutations to evaluate if these offer additional diagnostic accuracy for LNM detection. Nonetheless, our study provides an important proof for detecting LNM in patients with T1 CRC, and these findings are potentially an important major step toward the availability of robust molecular biomarkers for the risk assessment and management of a lethal malignancy.
In conclusion, we have identified and developed a novel risk-stratification model that allows identification of LNM in a liquid biopsy assay for more robust and accurate identification of patients with high-risk T1 CRC. Pending validation in future prospective studies, our findings highlight the potential clinical impact of our model for improved selection of patients with high-risk T1 CRC, which will reduce the overall burden of unnecessary surgeries and expense associated with these procedures, and improve the overall management of patients with this malignancy.
Supplementary Material
Supplemental Figure S1. ROC curves for the detection of LNM in T1 CRC training and validation cohorts. A) ROC curve for combined current clinical risk factors (depth of submucosal invasion (>1000 μm), presence of lymphatic or vascular invasion, high-grade tumor budding, and poorly differentiated histology) for LNM without the transcriptomic panel in the training cohort (AUC = 0.73). B) ROC curve for the current clinical risk factors for LNM without the transcriptomic panel in the validation cohort (AUC = 0.76).
Table 1.
Clinicopathologic Characteristics of Clinical Cohorts
| Characteristics | Training cohort (N = 46) | Validation cohort (N = 142) | P |
|---|---|---|---|
|
| |||
| Age, y | |||
| Median (range) | 70 (38–90) | 67 (24–85) | .16 |
| Sex | |||
| Male | 24 (52) | 86 (61) | |
| Female | 22 (48) | 56 (39) | .32 |
| LNM | |||
| Positive | 5 (11) | 12 (8) | |
| Negative | 41 (89) | 130 (92) | .62 |
| MSI status | |||
| MSI-H | 10 (7) | ||
| MSI-L | 5 (4) | ||
| MSS | 127 (89) | ... | |
| Tumor location | |||
| Right side | 17 (37) | 41 (29) | |
| Left side | 29 (63) | 101 (71) | .3 |
| Tumor size, mm | |||
| ≥20 | 23 (50) | 65 (46) | |
| <20 | 23 (50) | 77 (54) | .18 |
| Submucosal invasion, mm | |||
| ≥1000 | 43 (93) | 140 (99) | |
| <1000 | 3 (7) | 2 (1) | .16 |
| Budding grade | |||
| ≥2 | 9 (20) | 26 (18) | |
| 1 | 27 (58) | 104 (74) | |
| Unavailable | 10 (22) | 12 (8) | .52 |
| Lymph invasion | |||
| Positive | 13 (28) | 53 (37) | |
| Negative | 33 (72) | 89 (63) | .26 |
| Vascular invasion | |||
| Positive | 13 (28) | 49 (35) | |
| Negative | 33 (72) | 93 (65) | .43 |
| Differentiation | |||
| Well | 22 (48) | 97 (68) | |
| Moderate | 21 (46) | 43 (31) | |
| Poor | 2 (4) | 0 (0) | |
NOTE. Data are shown as n (%) unless indicated otherwise.
MSI, microsatellite instability; MSI-H, high-frequency microsatellite instability; MSI-L, low-frequency microsatellite instability; MSS, microsatellite stable.
WHAT YOU NEED TO KNOW.
BACKGROUND AND CONTEXT
The patients with high-risk invasive submucosal colorectal cancers (T1 CRCs) are often recommended radical surgery due to the risk of lymph node metastases (LNM), but the current pathological criteria for risk-stratification of LNM are inadequate and often lead to overtreatment.
NEW FINDINGS
Using serum and matched tumor specimens from independent patient cohorts, we herein report a noninvasive, liquid biopsy transcriptomic assay that can robustly identify patients at risk for LNM prior to surgery. By combining this biomarker panel with key clinical features, we established a risk-stratification model that exhibited superior accuracy for identification of LNM.
LIMITATIONS
This was a retrospective study, and independent prospective studies are needed to further confirm the diagnostic potential of this diagnostic assay prior to its translation in the clinic.
IMPACT
Our risk-stratification model has a potential to serve as a noninvasive, liquid biopsy assay to identify high-risk T1 CRC patients with LNM prior to surgery, and reduce the overall burden of unnecessary surgeries and expense associated with these procedures.
ACKNOWLEDGEMENTS
We would like to thank Drs. Raju Kandimalla, Tatsuhiko Kakisaka, Satoshi Nishiwada, Yasuyuki Okada, Huanlin Wang, In-Seob Lee, Divya Sahu, Geeta Sharma, and Souvick Roy for their thoughtful discussions and advice during the course of this project. We also would like to extend our thanks to Dr. Sarah Wilkinson for her significant editing and useful suggestions for improving the quality of our manuscript. The serum samples used in this study were partially obtained from the National Cancer Center Biobank, which is supported by the National Cancer Center Research and Development Fund (29-A-1).
Funding:
This work was supported by CA72851, CA181572, CA184792, CA202797, and CA227602 grants from the National Cancer Institute, National Institutes of Health.
Glossary
- RT-qPCR
Real-time quantitative reverse transcription polymerase chain reaction
- T1 CRC
Invasive submucosal colorectal cancer
Footnotes
Conflict of Interest: None of the authors has any potential conflicts to disclose.
CRediT Authorship Contributions:
Yuma Wada (Conceptualization: Equal; Data curation: Lead; Formal analysis: Lead; Methodology: Lead; Validation: Lead; Writing - original draft: Lead; Writing - review & editing: Equal). Mitsuo Shimada (Formal analysis: Supporting; Writing - review & editing: Supporting). Tatsuro Murano (Resources: Equal; Validation: Supporting; Writing - review & editing: Equal). Hiroyuki Takamaru (Resources: Equal; Validation: Supporting; Writing - review & editing: Supporting). Yuji Morine (Data curation: Supporting; Validation: Supporting; Writing - review & editing: Supporting). Tetsuya Ikemoto (Formal analysis: Supporting; Writing - review & editing: Supporting). Yu Saito (Methodology: Supporting; Writing - review & editing: Supporting). Francesc Balaguer (Conceptualization: Supporting; Resources: Supporting; Writing - review & editing: Equal). Luis Bujanda (Resources: Supporting; Validation: Supporting; Writing - review & editing: Supporting). Maria Pellise (Resources: Supporting; Writing - review & editing: Supporting). Ken Kato (Resources: Equal). Yutaka Saito (Resources: Equal; Writing - review & editing: Supporting). Hiroaki Ikematsu (Resources: Equal; Writing - review & editing: Equal). Ajay Goel (Conceptualization: Lead; Funding acquisition: Lead; Writing - review & editing: Lead).
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Stamos MJ, Murrell Z. Management of early rectal T1 and T2 cancers. Clin Cancer Res 2007;13:6885s–9s. [DOI] [PubMed] [Google Scholar]
- 2.Iida S, Hasegawa H, Okabayashi K, et al. Risk factors for postoperative recurrence in patients with pathologically T1 colorectal cancer. World J Surg 2012;36:424–30. [DOI] [PubMed] [Google Scholar]
- 3.Tanaka S, Kashida H, Saito Y, et al. JGES guidelines for colorectal endoscopic submucosal dissection/endoscopic mucosal resection. Dig Endosc 2015;27:417–434. [DOI] [PubMed] [Google Scholar]
- 4.Shimura T, Ebi M, Yamada T, et al. Magnifying chromoendoscopy and endoscopic ultrasonography measure invasion depth of early stage colorectal cancer with equal accuracy on the basis of a prospective trial. Clin Gastroenterol Hepatol 2014;12:662–8 e1–2. [DOI] [PubMed] [Google Scholar]
- 5.Vleugels JLA, Koens L, Dijkgraaf MGW, et al. Suboptimal endoscopic cancer recognition in colorectal lesions in a national bowel screening programme. Gut 2020;69:977–980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Muto T, Oya M. Recent advances in diagnosis and treatment of colorectal T1 carcinoma. Dis Colon Rectum 2003;46:S89–93. [DOI] [PubMed] [Google Scholar]
- 7.Tanaka S, Haruma K, Oh EH, et al. Conditions of curability after endoscopic resection for colorectal carcinoma with submucosally massive invasion. Oncol Rep 2000;7:783–8. [DOI] [PubMed] [Google Scholar]
- 8.Nascimbeni R, Burgart LJ, Nivatvongs S, et al. Risk of lymph node metastasis in T1 carcinoma of the colon and rectum. Dis Colon Rectum 2002;45:200–6. [DOI] [PubMed] [Google Scholar]
- 9.Sohn DK, Chang HJ, Park JW, et al. Histopathological risk factors for lymph node metastasis in submucosal invasive colorectal carcinoma of pedunculated or semipedunculated type. J Clin Pathol 2007;60:912–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Suh JH, Han KS, Kim BC, et al. Predictors for lymph node metastasis in T1 colorectal cancer. Endoscopy 2012;44:590–5. [DOI] [PubMed] [Google Scholar]
- 11.Watanabe T, Itabashi M, Shimada Y, et al. Japanese Society for Cancer of the Colon and Rectum (JSCCR) Guidelines 2014 for treatment of colorectal cancer. Int J Clin Oncol 2015;20:207–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ueno H, Mochizuki H, Hashiguchi Y, et al. Risk factors for an adverse outcome in early invasive colorectal carcinoma. Gastroenterology 2004;127:385–94. [DOI] [PubMed] [Google Scholar]
- 13.Choi DH, Sohn DK, Chang HJ, et al. Indications for subsequent surgery after endoscopic resection of submucosally invasive colorectal carcinomas: a prospective cohort study. Dis Colon Rectum 2009;52:438–45. [DOI] [PubMed] [Google Scholar]
- 14.Tateishi Y, Nakanishi Y, Taniguchi H, et al. Pathological prognostic factors predicting lymph node metastasis in submucosal invasive (T1) colorectal carcinoma. Mod Pathol 2010;23:1068–72. [DOI] [PubMed] [Google Scholar]
- 15.Ikematsu H, Yoda Y, Matsuda T, et al. Long-term outcomes after resection for submucosal invasive colorectal cancers. Gastroenterology 2013;144:551–9; quiz e14. [DOI] [PubMed] [Google Scholar]
- 16.Yoda Y, Ikematsu H, Matsuda T, et al. A large-scale multicenter study of long-term outcomes after endoscopic resection for submucosal invasive colorectal cancer. Endoscopy 2013;45:718–24. [DOI] [PubMed] [Google Scholar]
- 17.Nakadoi K, Tanaka S, Kanao H, et al. Management of T1 colorectal carcinoma with special reference to criteria for curative endoscopic resection. J Gastroenterol Hepatol 2012;27:1057–62. [DOI] [PubMed] [Google Scholar]
- 18.Pimentel-Nunes P, Dinis-Ribeiro M, Ponchon T, et al. Endoscopic submucosal dissection: European Society of Gastrointestinal Endoscopy (ESGE) Guideline. Endoscopy 2015;47:829–54. [DOI] [PubMed] [Google Scholar]
- 19.Ha RK, Han KS, Sohn DK, et al. Histopathologic risk factors for lymph node metastasis in patients with T1 colorectal cancer. Ann Surg Treat Res 2017;93:266–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yamamoto S, Watanabe M, Hasegawa H, et al. The risk of lymph node metastasis in T1 colorectal carcinoma. Hepatogastroenterology 2004;51:998–1000. [PubMed] [Google Scholar]
- 21.Son HJ, Song SY, Lee WY, et al. Characteristics of early colorectal carcinoma with lymph node metastatic disease. Hepatogastroenterology 2008;55:1293–7. [PubMed] [Google Scholar]
- 22.Okabe S, Shia J, Nash G, et al. Lymph node metastasis in T1 adenocarcinoma of the colon and rectum. J Gastrointest Surg 2004;8:1032–9; discussion 1039–40. [DOI] [PubMed] [Google Scholar]
- 23.Tamaru Y, Oka S, Tanaka S, et al. Long-term outcomes after treatment for T1 colorectal carcinoma: a multicenter retrospective cohort study of Hiroshima GI Endoscopy Research Group. J Gastroenterol 2017;52:1169–1179. [DOI] [PubMed] [Google Scholar]
- 24.Ricciardi R, Madoff RD, Rothenberger DA, et al. Population-based analyses of lymph node metastases in colorectal cancer. Clin Gastroenterol Hepatol 2006;4:1522–7. [DOI] [PubMed] [Google Scholar]
- 25.Brunner W, Widmann B, Marti L, et al. Predictors for regional lymph node metastasis in T1 rectal cancer: a population-based SEER analysis. Surg Endosc 2016;30:4405–15. [DOI] [PubMed] [Google Scholar]
- 26.Nozawa H, Ishihara S, Fujishiro M, et al. Outcome of salvage surgery for colorectal cancer initially treated by upfront endoscopic therapy. Surgery 2016;159:713–20. [DOI] [PubMed] [Google Scholar]
- 27.Chen X, Ba Y, Ma L, et al. Characterization of microRNAs in serum: a novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res 2008;18:997–1006. [DOI] [PubMed] [Google Scholar]
- 28.Huang Z, Huang D, Ni S, et al. Plasma microRNAs are promising novel biomarkers for early detection of colorectal cancer. Int J Cancer 2010;127:118–26. [DOI] [PubMed] [Google Scholar]
- 29.Kanaan Z, Rai SN, Eichenberger MR, et al. Plasma miR-21: a potential diagnostic marker of colorectal cancer. Ann Surg 2012;256:544–51. [DOI] [PubMed] [Google Scholar]
- 30.Ng EK, Chong WW, Jin H, et al. Differential expression of microRNAs in plasma of patients with colorectal cancer: a potential marker for colorectal cancer screening. Gut 2009;58:1375–81. [DOI] [PubMed] [Google Scholar]
- 31.Toiyama Y, Takahashi M, Hur K, et al. Serum miR-21 as a diagnostic and prognostic biomarker in colorectal cancer. J Natl Cancer Inst 2013;105:849–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hur K, Toiyama Y, Okugawa Y, et al. Circulating microRNA-203 predicts prognosis and metastasis in human colorectal cancer. Gut 2017;66:654–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Toiyama Y, Hur K, Tanaka K, et al. Serum miR-200c is a novel prognostic and metastasis-predictive biomarker in patients with colorectal cancer. Ann Surg 2014;259:735–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ozawa T, Kandimalla R, Gao F, et al. A MicroRNA Signature Associated With Metastasis of T1 Colorectal Cancers to Lymph Nodes. Gastroenterology 2018;154:844–848.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kandimalla R, Ozawa T, Gao F, et al. Gene Expression Signature in Surgical Tissues and Endoscopic Biopsies Identifies High-Risk T1 Colorectal Cancers. Gastroenterology 2019;156:2338–2341.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001;25:402–8. [DOI] [PubMed] [Google Scholar]
- 37.Jepsen RK, Novotny GW, Klarskov LL, et al. Intra-tumor heterogeneity of microRNA-92a, microRNA-375 and microRNA-424 in colorectal cancer. Exp Mol Pathol 2016;100:125–31. [DOI] [PubMed] [Google Scholar]
- 38.Studdert DM, Mello MM, Sage WM, et al. Defensive medicine among high-risk specialist physicians in a volatile malpractice environment. Jama 2005;293:2609–17. [DOI] [PubMed] [Google Scholar]
- 39.Eamer GJ, Clement F, Pederson JL, et al. Analysis of postdischarge costs following emergent general surgery in elderly patients. Can J Surg 2018;61:19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kim B, Kim EH, Park SJ, et al. The risk of lymph node metastasis makes it unsafe to expand the conventional indications for endoscopic treatment of T1 colorectal cancer: A retrospective study of 428 patients. Medicine (Baltimore) 2016;95:e4373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Overwater A, Kessels K, Elias SG, et al. Endoscopic resection of high-risk T1 colorectal carcinoma prior to surgical resection has no adverse effect on long-term outcomes. Gut 2018;67:284–290. [DOI] [PubMed] [Google Scholar]
- 42.Barel F, Cariou M, Saliou P, et al. Histopathological factors help to predict lymph node metastases more efficiently than extra-nodal recurrences in submucosa invading pT1 colorectal cancer. Sci Rep 2019;9:8342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Imaoka H, Toiyama Y, Fujikawa H, et al. Circulating microRNA-1290 as a novel diagnostic and prognostic biomarker in human colorectal cancer. Ann Oncol 2016;27:1879–86. [DOI] [PubMed] [Google Scholar]
- 44.Kishida Y, Oishi T, Sugino T, et al. Associations Between Loss of ARID1A Expression and Clinicopathologic and Genetic Variables in T1 Early Colorectal Cancer. Am J Clin Pathol 2019;152:463–470. [DOI] [PubMed] [Google Scholar]
- 45.Haasnoot KJC, Backes Y, Moons LMG, et al. Associations of non-pedunculated T1 colorectal adenocarcinoma outcome with consensus molecular subtypes, immunoscore, and microsatellite status: a multicenter case-cohort study. Mod Pathol 2020;33:2626–2636. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure S1. ROC curves for the detection of LNM in T1 CRC training and validation cohorts. A) ROC curve for combined current clinical risk factors (depth of submucosal invasion (>1000 μm), presence of lymphatic or vascular invasion, high-grade tumor budding, and poorly differentiated histology) for LNM without the transcriptomic panel in the training cohort (AUC = 0.73). B) ROC curve for the current clinical risk factors for LNM without the transcriptomic panel in the validation cohort (AUC = 0.76).


