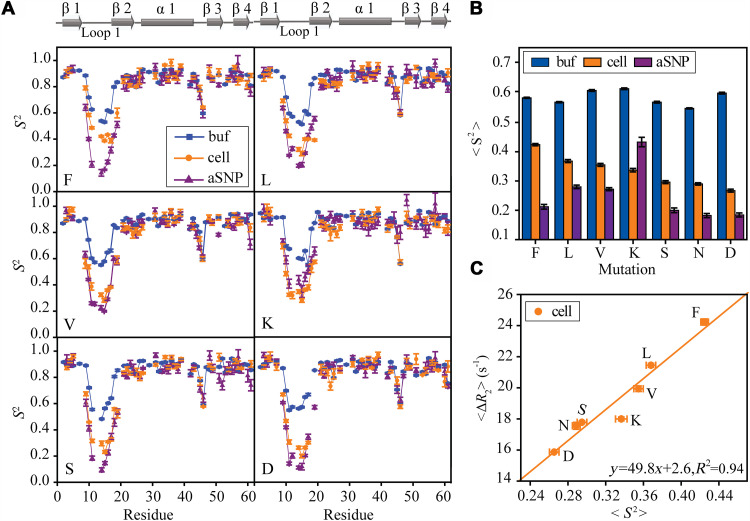
Fig. 5. GB3L conformational dynamics and its sequence dependence.
The loop 1 sequence was varied by changing X in GXSGG to different amino acids. (A) S2 order parameters for different mutants, F, L, V, K, S, and D. (B) Averages of loop S2 (including G11, G14, G15, T16, and L17, which have S2 values in all GB3L variants) of different mutants in the buffer only, in the presence of anionic SNP, and in cells. The loop S2 average in cells (C) is correlated with <ΔR2> [ΔR2 = R2(cell) − R2(buffer)] averaged over rigid GB3L residues. Error bars are SEM (n = 3 independent experiments).