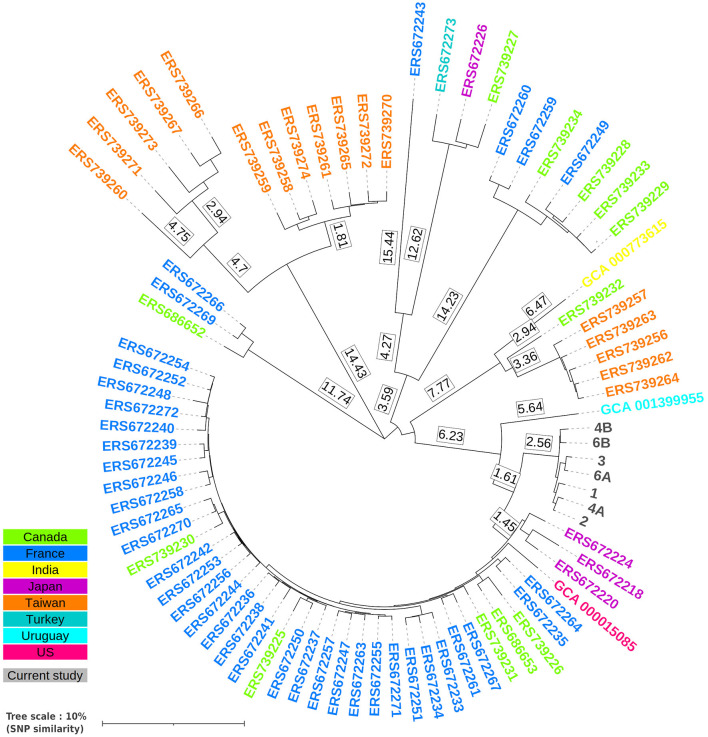
Figure 1.
Whole-genome SNP phylogenetic tree generated from the alignment of each C. fetus studied isolate (cases 1, 2, 3, 4A, 4B, 6A, and 6B) combined with 75 additional clinical isolates (Iraola et al., 2017) from various countries against C. fetus reference CFF00A031. Genomes were aligned against the reference using bwa mem, and SNPs were called using samtools. Pair-wise comparison between all genomes was performed in order to determine the proportion of genotype similarity, which is displayed in the present tree in the percentage of identical SNP using iTOL online tool v6.