Figure 2. Simulations of subunit rotation in a eukaryotic ribosome.
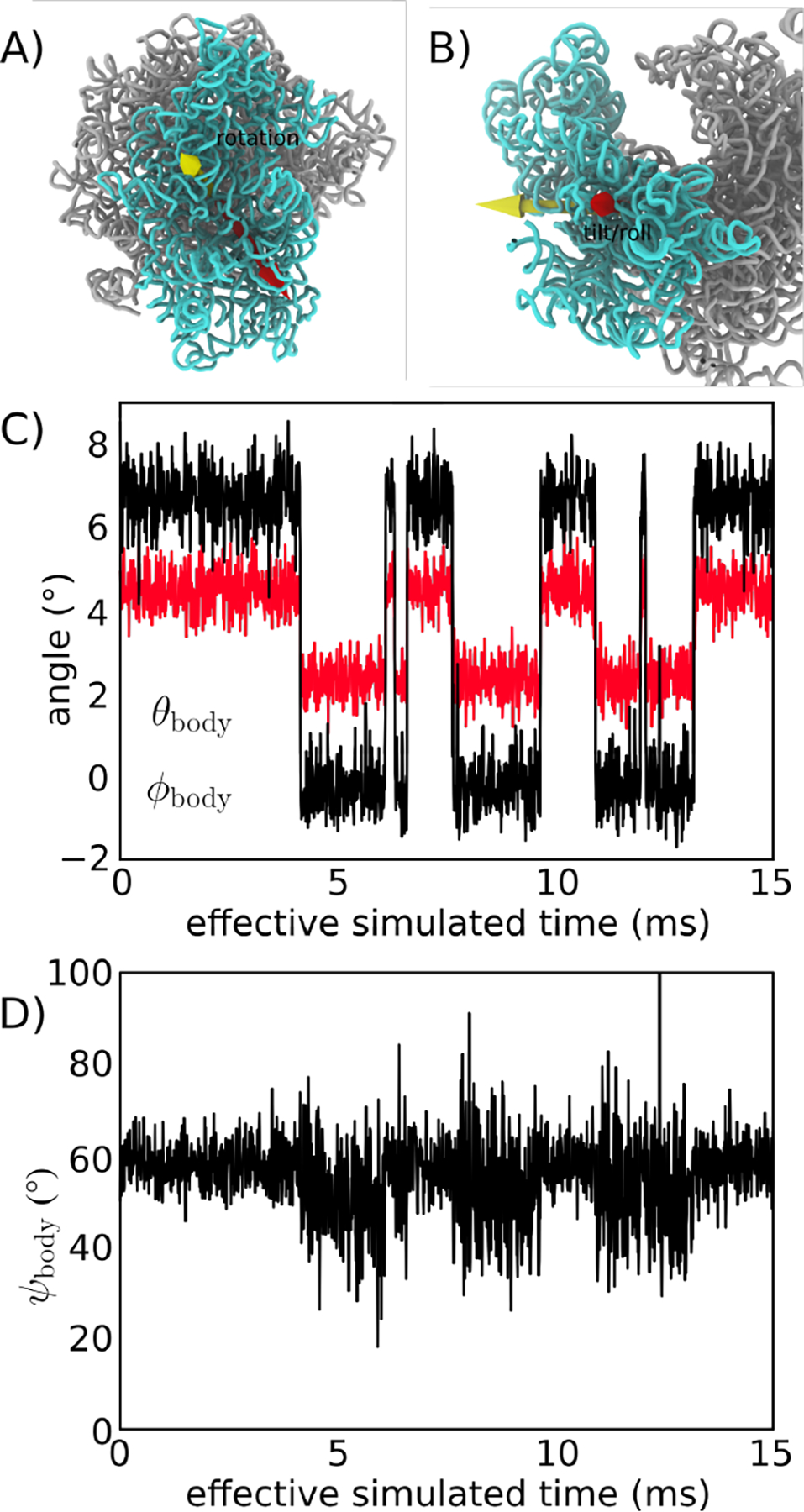
Using an all-atom structure-based model [24], we simulated spontaneous rotation and backrotation of the SSU in yeast. A) Euler angle decomposition was used to describe the orientation of the SSU, where the angle measures rotation of the SSU body, relative to the LSU. is defined as rotation about the yellow vector. B) The tile angle describes rotation that is orthogonal to the body rotation angle . The tilt axis (red) may be in any direction perpendicular to the rotation axis (yellow), where the direction of tilting axis is given by . In the figure, is shown. C) and shown for a single simulation (of 3, in total). In this model, there are distinct and sharp transitions between the unrotated and rotated orientations. There are also concommittent changes in . For reference, the cryo-EM structures [14] correspond to and for the unrotated state and and for the rotated state. Negative and non-zero angles for the unrotated conformation reflect the relative orientation of the SSU in yeast, relative to the reference bacterial system (E. coli). D) In the simulation, the direction of tilting shifts to slightly higher values as the SSU rotates and tilts. This reveals how the direction of structural fluctuations depends on the global conformation of the ribosome. The effective simulated times are estimated based on previous comparisons with explicit-solvent simulations [31]
