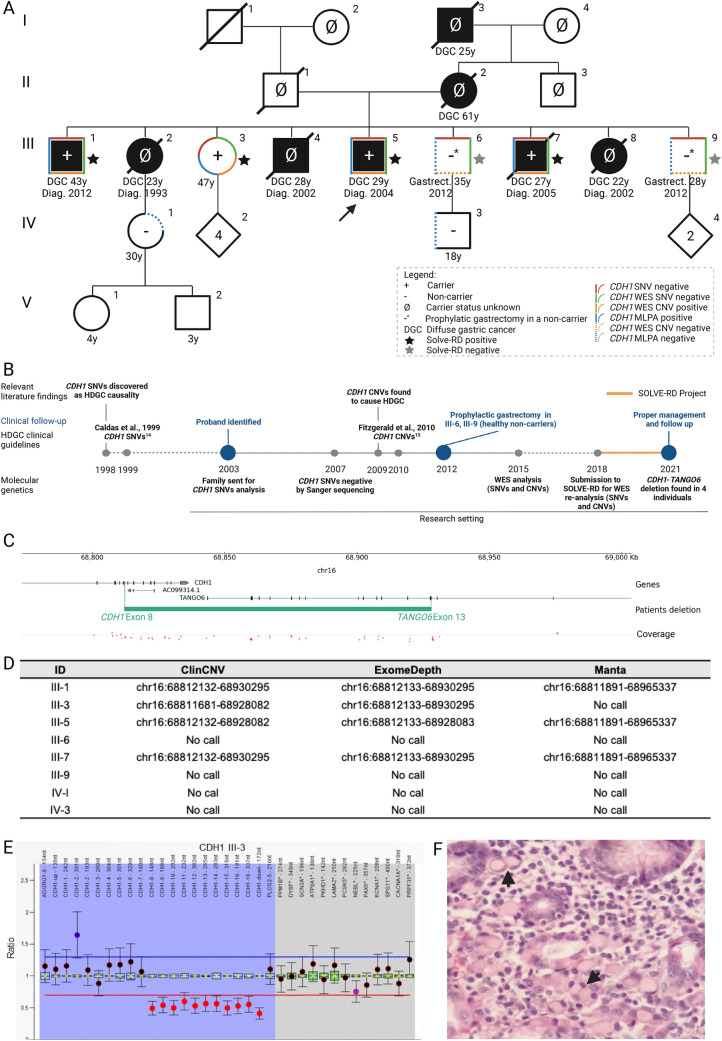
Fig. 1.
Diagnostic odyssey, pedigree, variant calling details and cancer histology of a HDGC family carrying a causative CDH1 CNV. A Current family pedigree. Full black symbols: individuals with confirmed DGC; Red outline: negative for SNVs (Sanger sequencing); green outline: negative for SNVs (WES); orange outline: positive for CNVs (WES); blue outline: MLPA-positive for the CDH1 deletion; traced orange outline: negative for CNVs (WES); traced blue outline: MLPA-negative for the CDH1 deletion. All family members, submitted to multiple genetic tests herein described, signed informed consents. The research work has been approved by the Ethics Committee of Centro Hospitalar Universitário São João, in Porto, Portugal, in the frame of the Solve-RD project with the reference CHUSJ_445/2020; B Timeline of events and diagnostic odyssey of the family; C CDH1 CNV found by WES, encompassing part of the CDH1 and TANGO6 genes (represented in green) and IGV coverage; Green text represents first and last edited exons; D Detected CDH1 CNV in ClinCNV [31], ExomeDepth [30] and Manta [32]; E MLPA analysis performed using SALSA MLPA-Probemix P083 CDH1 (MRC Holland) in patient III-3; Ratio above blue line indicates increased copy number, whilst ratio below red line indicates reduced copy number; Blue highlight represents CDH1 probes and 2 neighbour genes; Grey highlight represents reference probes; F Haematoxylin and eosin staining of DGC depicting signet-ring cells (arrow heads) (100 × magnification) from the proband III-5