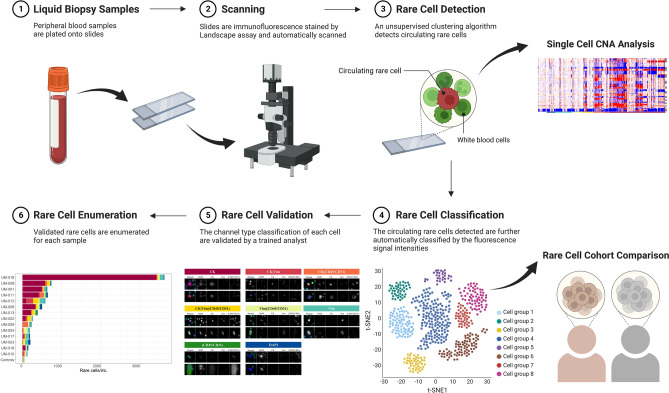
Figure 1.
HDSCA3.0 Workflow. (1) Peripheral blood samples are plated onto slides. (2) Slides are immunofluorescence stained by Landscape assay and automatically scanned. (3) Detection of rare cells from the imaging data set is conducted using an unsupervised clustering algorithm that clusters rare cells using extracted quantitative morphologic features. Further downstream analysis of single-cell copy number alteration (CNA) on those detected rare cells is performed. (4) The circulating rare cells detected are further automatically classified into channel-based cell classifications defined by the fluorescence signal intensities. (5) The channel-based cell classification of each cell is validated by a trained analyst. (6) Enumeration of each rare cell type per sample is counted.