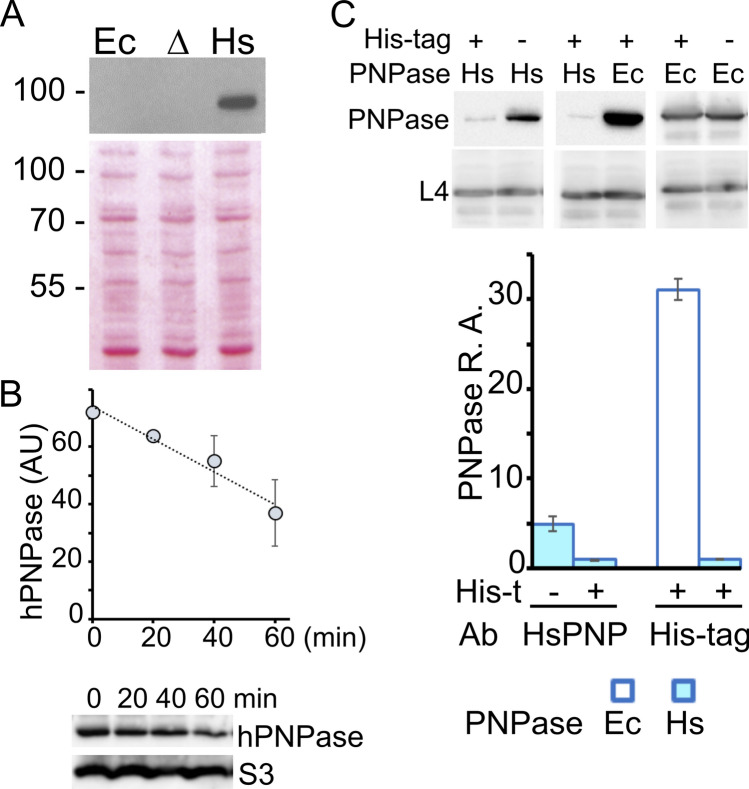
Figure 3.
Abundance and stability of hPNPase in the PNPT1Ec strain. (A) Western blotting of proteins extracted from exponential cultures of C-1a (pnp+; Ec), C-5691 (Δpnp; Δ) and C-6001 (PNPT1Ec; Hs). The proteins (10 µg) were run on 10% polyacrylamide-SDS gel and blotted onto a nitrocellulose membrane. The filter was stained with Ponceau S (lower panel) to check loading and hybridized with hPNPase-specific monoclonal antibodies (upper panel). The position of MW markers is reported on the left in kDa. (B) hPNPase stability. 15 µl of PNPT1Ec strain protein extracts (0.375 OD600) were loaded onto 10% polyacrylamide gel, blotted onto a nitrocellulose membrane and hybridized with polyclonal anti-hPNPase and anti-S3 antibodies. The results of a typical experiment are shown under the graph. To plot the decay curve, hPNPase signals were quantified with ImageLab (Bio-Rad), using S3 signals as loading control. Symbols in the plot represent mean with StD (N = 3). AU, arbitrary units. (C) Upper part. Western blotting of proteins extracted from exponential cultures of C-6009 (+, Hs); C-6001 (−, Hs), C-6011 (+, Ec) and C-1a (−, Ec) grown at 37 °C in LD up to OD600 = 0.4. The proteins were run on a 12% polyacrylamide- SDS gel, blotted onto a nitrocellulose membrane and hybridized with hPNPase—(upper left panel), His tag—(upper central panel), EcPNPase—(upper right panel) or L4—specific (lower panels, loading control) antibodies. Lower part, PNPase Relative Amount (R. A.). The bars represent average with StD (N = 3) of quantification of PNPase signals with Image Lab (Bio-Rad).