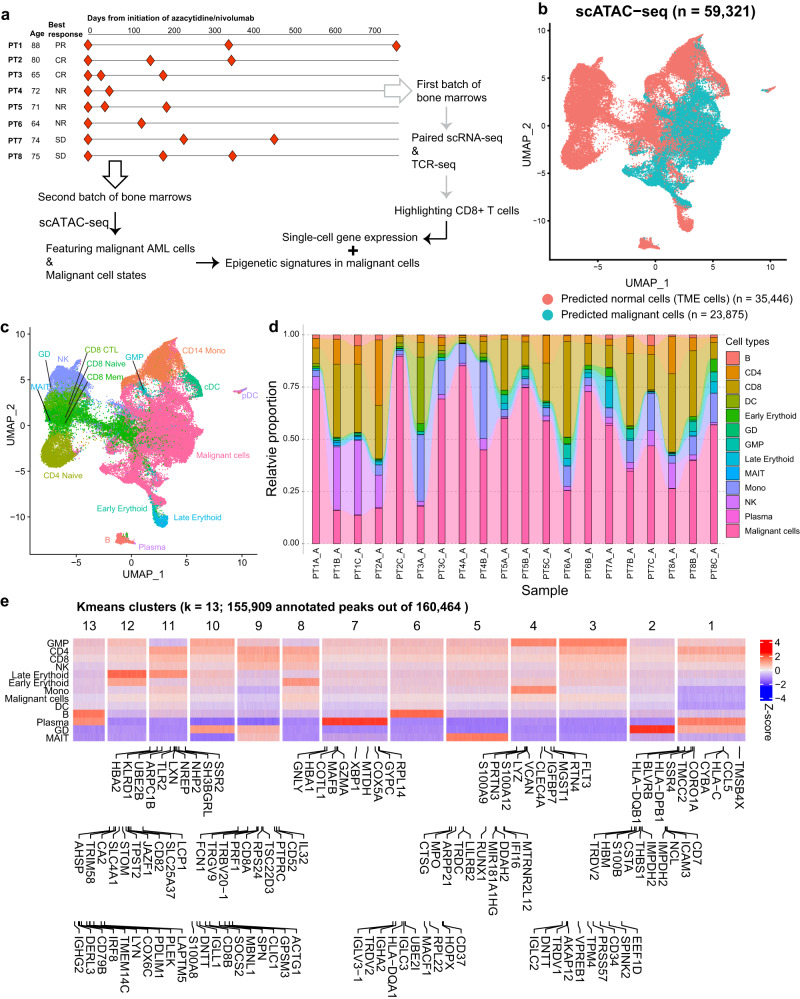
Fig. 1. Open-chromatin landscape in AML patients profiled using scATAC-seq.
a An introduction of the recruited cohort for our study which included eight patients. Longitudinal bone marrow samples were extracted per patient for sequencing. First batch of samples were profiled using paired scRNA-seq and TCR-seq, with a highlight of CD8 T cells (published), while second batch of samples were profiled using scATAC-seq and centered on the analyses of malignant cells by including gene expression as a complement. b UMAP of scATAC-seq clusters colored by tumor microenvironment (TME) and malignant cells. c UMAP of scATAC-seq clusters colored and labeled by granular cell types. d Barchart showing relative proportions of predicted cell types across different samples. e Heatmap showing all annotated peaks (n = 155,909) aggregated by cell types. Each column represents a peak region, while each row represents a cell type. Peaks are split into 13 K-means clusters, with top ten differentially expressed genes (calculated using our published scRNA-seq data) labeled along each cluster.