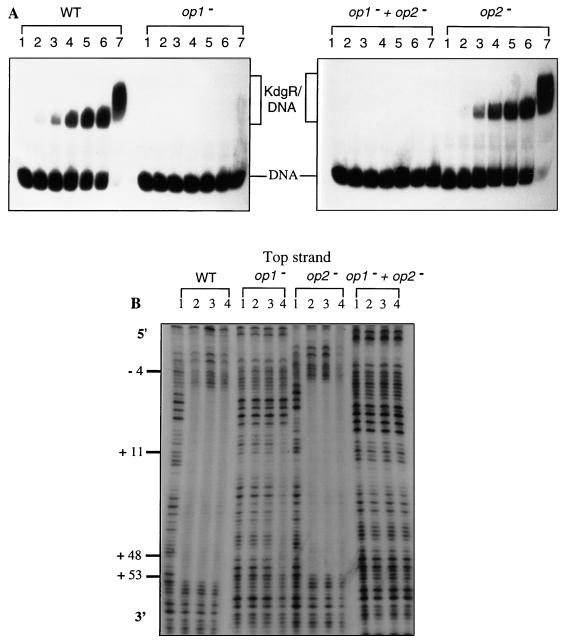
FIG. 4.
Interactions between the KdgR repressor and the wild-type E. chrysanthemi EC16 pelE operator (WT) or its derivatives modified in OP1 (op1−), in OP2 (op2−), or in both (op1− + op2−). (A) Band shift assay. The DNA fragment (about 10 fmol), isolated and labeled as described in Materials and Methods, was incubated with 0, 1, 5, 10, 15, 20, or 200 nM purified KdgR (lane 1, 2, 3, 4, 5, 6, or 7, respectively). (B) DNase I footprinting experiments. The purified KdgR protein was added to a final concentration of 20, 75, or 200 nM (lane 2, 3, or 4, respectively). Lanes 1, control digestion. The sequences are numbered with respect to the transcription start site (+1).